Risk Prediction Using Genome-Wide Association Studies on Type 2 Diabetes
- PMID: 28154504
- PMCID: PMC5287117
- DOI: 10.5808/GI.2016.14.4.138
Risk Prediction Using Genome-Wide Association Studies on Type 2 Diabetes
Abstract
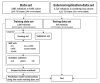
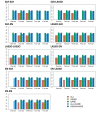
The success of genome-wide association studies (GWASs) has enabled us to improve risk assessment and provide novel genetic variants for diagnosis, prevention, and treatment. However, most variants discovered by GWASs have been reported to have very small effect sizes on complex human diseases, which has been a big hurdle in building risk prediction models. Recently, many statistical approaches based on penalized regression have been developed to solve the "large p and small n" problem. In this report, we evaluated the performance of several statistical methods for predicting a binary trait: stepwise logistic regression (SLR), least absolute shrinkage and selection operator (LASSO), and Elastic-Net (EN). We first built a prediction model by combining variable selection and prediction methods for type 2 diabetes using Affymetrix Genome-Wide Human SNP Array 5.0 from the Korean Association Resource project. We assessed the risk prediction performance using area under the receiver operating characteristic curve (AUC) for the internal and external validation datasets. In the internal validation, SLR-LASSO and SLR-EN tended to yield more accurate predictions than other combinations. During the external validation, the SLR-SLR and SLR-EN combinations achieved the highest AUC of 0.726. We propose these combinations as a potentially powerful risk prediction model for type 2 diabetes.
Keywords: clinical prediction rule; genome-wide association study; penalized regression models; type 2 diabetes mellitus.
Figures


References
-
- Wang WY, Barratt BJ, Clayton DG, Todd JA. Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet. 2005;6:109–118. - PubMed
-
- Evans DM, Visscher PM, Wray NR. Harnessing the information contained within genome-wide association studies to improve individual prediction of complex disease risk. Hum Mol Genet. 2009;18:3525–3531. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
