LC-MS-Based Metabolic Fingerprinting of Aqueous Humor
- PMID: 28154769
- PMCID: PMC5244013
- DOI: 10.1155/2017/6745932
LC-MS-Based Metabolic Fingerprinting of Aqueous Humor
Abstract
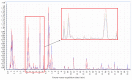
Aqueous humor (AH) is a transparent fluid which fills the anterior and posterior chambers of the eye. It supplies nutrients and removes metabolic waste from avascular tissues in the eye. Proper homeostasis of AH is required to maintain adequate intraocular pressure as well as optical and refractive properties of the eye. Application of metabolomics to study human AH may improve knowledge about the molecular mechanisms of eye diseases. Until now, global analysis of metabolites in AH has been mainly performed using NMR. Among the analytical platforms used in metabolomics, LC-MS allows for the highest metabolome coverage. The aim of this study was to develop a method for extraction and analysis of AH metabolites by LC-QTOF-MS. Different protocols for AH preparation were tested. The best results were obtained when one volume of AH was mixed with one volume of methanol : ethanol (1 : 1). In the final method, 2 µL of extracted sample was analyzed by LC-QTOF-MS. The method allowed for reproducible measurement of over 1000 metabolic features. Almost 250 metabolites were identified in AH and assigned to 47 metabolic pathways. This method is suitable to study the potential role of amino acids, lipids, oxidative stress, or microbial metabolites in development of ocular diseases.
Conflict of interest statement
The authors declare that there is no conflict of interests regarding the publication of this manuscript.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources

