A study on the application of topic models to motif finding algorithms
- PMID: 28155646
- PMCID: PMC5259985
- DOI: 10.1186/s12859-016-1364-3
A study on the application of topic models to motif finding algorithms
Abstract
Background: Topic models are statistical algorithms which try to discover the structure of a set of documents according to the abstract topics contained in them. Here we try to apply this approach to the discovery of the structure of the transcription factor binding sites (TFBS) contained in a set of biological sequences, which is a fundamental problem in molecular biology research for the understanding of transcriptional regulation. Here we present two methods that make use of topic models for motif finding. First, we developed an algorithm in which first a set of biological sequences are treated as text documents, and the k-mers contained in them as words, to then build a correlated topic model (CTM) and iteratively reduce its perplexity. We also used the perplexity measurement of CTMs to improve our previous algorithm based on a genetic algorithm and several statistical coefficients.
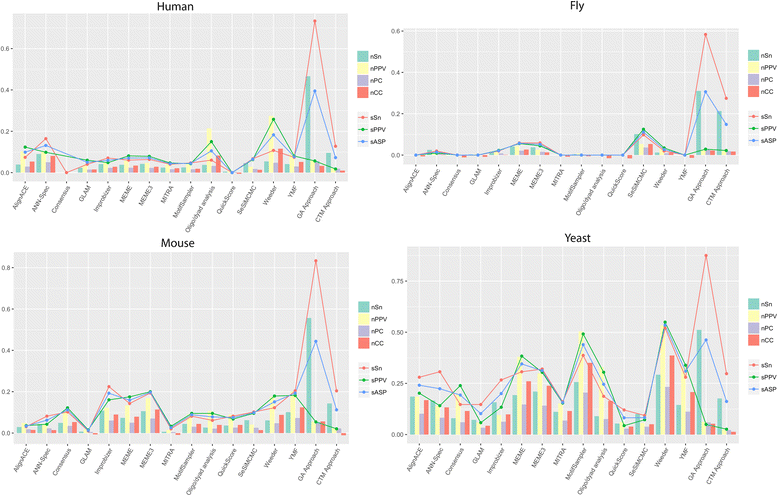
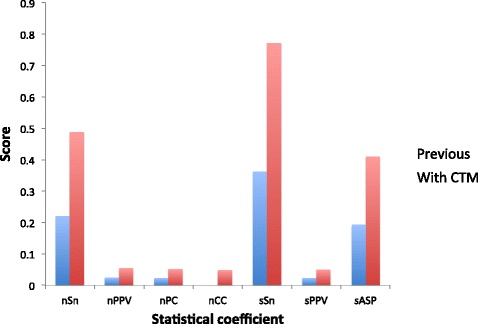
Results: The algorithms were tested with 56 data sets from four different species and compared to 14 other methods by the use of several coefficients both at nucleotide and site level. The results of our first approach showed a performance comparable to the other methods studied, especially at site level and in sensitivity scores, in which it scored better than any of the 14 existing tools. In the case of our previous algorithm, the new approach with the addition of the perplexity measurement clearly outperformed all of the other methods in sensitivity, both at nucleotide and site level, and in overall performance at site level.
Conclusions: The statistics obtained show that the performance of a motif finding method based on the use of a CTM is satisfying enough to conclude that the application of topic models is a valid method for developing motif finding algorithms. Moreover, the addition of topic models to a previously developed method dramatically increased its performance, suggesting that this combined algorithm can be a useful tool to successfully predict motifs in different kinds of sets of DNA sequences.
Figures



References
-
- Blei DM. Probabilistic topic models. Commun. ACM. 2012;55(4):77–84. doi: 10.1145/2133806.2133826. - DOI
-
- Blei DM, Ng AY, Jordan MI. Latent dirichlet allocation. J. Mach. Learn. Res. 2003;3(Jan):993–1022.
-
- Gutierrez JB, Frith M, Nakai K. A Genetic Algorithm for Motif Finding Based on Statistical Significance. In: International Conference on Bioinformatics and Biomedical Engineering. Granada: Springer International Publishing; 2015. p. 438–49.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

