Epigenetic origin of evolutionary novel centromeres
- PMID: 28155877
- PMCID: PMC5290474
- DOI: 10.1038/srep41980
Epigenetic origin of evolutionary novel centromeres
Abstract
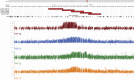
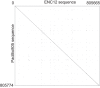
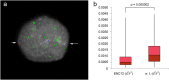
Most evolutionary new centromeres (ENC) are composed of large arrays of satellite DNA and surrounded by segmental duplications. However, the hypothesis is that ENCs are seeded in an anonymous sequence and only over time have acquired the complexity of "normal" centromeres. Up to now evidence to test this hypothesis was lacking. We recently discovered that the well-known polymorphism of orangutan chromosome 12 was due to the presence of an ENC. We sequenced the genome of an orangutan homozygous for the ENC, and we focused our analysis on the comparison of the ENC domain with respect to its wild type counterpart. No significant variations were found. This finding is the first clear evidence that ENC seedings are epigenetic in nature. The compaction of the ENC domain was found significantly higher than the corresponding WT region and, interestingly, the expression of the only gene embedded in the region was significantly repressed.
Conflict of interest statement
E.E.E. is on the scientific advisory board (SAB) of DNAnexus and is a consultant for Kunming University of Science and Technology (KUST) as part of the 1000 China Talent Program.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

