Microbes Are Associated with Host Innate Immune Response in Idiopathic Pulmonary Fibrosis
- PMID: 28157391
- PMCID: PMC5519968
- DOI: 10.1164/rccm.201607-1525OC
Microbes Are Associated with Host Innate Immune Response in Idiopathic Pulmonary Fibrosis
Abstract
Rationale: Differences in the lung microbial community influence idiopathic pulmonary fibrosis (IPF) progression. Whether the lung microbiome influences IPF host defense remains unknown.
Objectives: To explore the host immune response and microbial interaction in IPF as they relate to progression-free survival (PFS), fibroblast function, and leukocyte phenotypes.
Methods: Paired microarray gene expression data derived from peripheral blood mononuclear cells as well as 16S ribosomal RNA sequencing data from bronchoalveolar lavage obtained as part of the COMET-IPF (Correlating Outcomes with Biochemical Markers to Estimate Time-Progression in Idiopathic Pulmonary Fibrosis) study were used to conduct association pathway analyses. The responsiveness of paired lung fibroblasts to Toll-like receptor 9 (TLR9) stimulation by CpG-oligodeoxynucleotide (CpG-ODN) was integrated into microbiome-gene expression association analyses for a subset of individuals. The relationship between associated pathways and circulating leukocyte phenotypes was explored by flow cytometry.
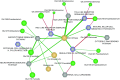
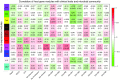
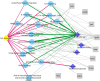
Measurements and main results: Down-regulation of immune response pathways, including nucleotide-binding oligomerization domain (NOD)-, Toll-, and RIG1-like receptor pathways, was associated with worse PFS. Ten of the 11 PFS-associated pathways correlated with microbial diversity and individual genus, with species accumulation curve richness as a hub. Higher species accumulation curve richness was significantly associated with inhibition of NODs and TLRs, whereas increased abundance of Streptococcus correlated with increased NOD-like receptor signaling. In a network analysis, expression of up-regulated signaling pathways was strongly associated with decreased abundance of operational taxonomic unit 1341 (OTU1341; Prevotella) among individuals with fibroblasts responsive to CpG-ODN stimulation. The expression of TLR signaling pathways was also linked to CpG-ODN responsive fibroblasts, OTU1341 (Prevotella), and Shannon index of microbial diversity in a network analysis. Lymphocytes expressing C-X-C chemokine receptor 3 CD8 significantly correlated with OTU1348 (Staphylococcus).
Conclusions: These findings suggest that host-microbiome interactions influence PFS and fibroblast responsiveness.
Keywords: CpG-oligodeoxynucleotide response; bronchoalveolar lavage microbiome; host immune response and microbial interaction; pattern recognition receptors; peripheral blood mononuclear cell transcriptome.
Figures

References
-
- Nathan SD, Shlobin OA, Weir N, Ahmad S, Kaldjob JM, Battle E, Sheridan MJ, du Bois RM. Long-term course and prognosis of idiopathic pulmonary fibrosis in the new millennium. Chest. 2011;140:221–229. - PubMed
-
- Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, Colby TV, Cordier JF, Flaherty KR, Lasky JA, et al. ATS/ERS/JRS/ALAT Committee on Idiopathic Pulmonary Fibrosis. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183:788–824. - PMC - PubMed
-
- Myers JL, Katzenstein AL. Epithelial necrosis and alveolar collapse in the pathogenesis of usual interstitial pneumonia. Chest. 1988;94:1309–1311. - PubMed
-
- Uhal BD, Joshi I, True AL, Mundle S, Raza A, Pardo A, Selman M. Fibroblasts isolated after fibrotic lung injury induce apoptosis of alveolar epithelial cells in vitro. Am J Physiol. 1995;269:L819–L828. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

