Dynamic Maternal Gradients Control Timing and Shift-Rates for Drosophila Gap Gene Expression
- PMID: 28158178
- PMCID: PMC5291410
- DOI: 10.1371/journal.pcbi.1005285
Dynamic Maternal Gradients Control Timing and Shift-Rates for Drosophila Gap Gene Expression
Erratum in
-
Correction: Dynamic Maternal Gradients Control Timing and Shift-Rates for Drosophila Gap Gene Expression.PLoS Comput Biol. 2017 Dec 22;13(12):e1005918. doi: 10.1371/journal.pcbi.1005918. eCollection 2017 Dec. PLoS Comput Biol. 2017. PMID: 29272271 Free PMC article.
Abstract
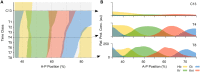
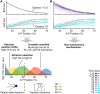
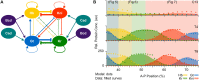
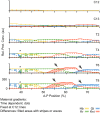
Pattern formation during development is a highly dynamic process. In spite of this, few experimental and modelling approaches take into account the explicit time-dependence of the rules governing regulatory systems. We address this problem by studying dynamic morphogen interpretation by the gap gene network in Drosophila melanogaster. Gap genes are involved in segment determination during early embryogenesis. They are activated by maternal morphogen gradients encoded by bicoid (bcd) and caudal (cad). These gradients decay at the same time-scale as the establishment of the antero-posterior gap gene pattern. We use a reverse-engineering approach, based on data-driven regulatory models called gene circuits, to isolate and characterise the explicitly time-dependent effects of changing morphogen concentrations on gap gene regulation. To achieve this, we simulate the system in the presence and absence of dynamic gradient decay. Comparison between these simulations reveals that maternal morphogen decay controls the timing and limits the rate of gap gene expression. In the anterior of the embyro, it affects peak expression and leads to the establishment of smooth spatial boundaries between gap domains. In the posterior of the embryo, it causes a progressive slow-down in the rate of gap domain shifts, which is necessary to correctly position domain boundaries and to stabilise the spatial gap gene expression pattern. We use a newly developed method for the analysis of transient dynamics in non-autonomous (time-variable) systems to understand the regulatory causes of these effects. By providing a rigorous mechanistic explanation for the role of maternal gradient decay in gap gene regulation, our study demonstrates that such analyses are feasible and reveal important aspects of dynamic gene regulation which would have been missed by a traditional steady-state approach. More generally, it highlights the importance of transient dynamics for understanding complex regulatory processes in development.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Pre-steady-state decoding of the Bicoid morphogen gradient.PLoS Biol. 2007 Feb;5(2):e46. doi: 10.1371/journal.pbio.0050046. PLoS Biol. 2007. PMID: 17298180 Free PMC article.
-
Bicoid determines sharp and precise target gene expression in the Drosophila embryo.Curr Biol. 2005 Nov 8;15(21):1888-98. doi: 10.1016/j.cub.2005.09.046. Curr Biol. 2005. PMID: 16271865
-
A system of repressor gradients spatially organizes the boundaries of Bicoid-dependent target genes.Cell. 2012 Apr 27;149(3):618-29. doi: 10.1016/j.cell.2012.03.018. Cell. 2012. PMID: 22541432 Free PMC article.
-
A matter of time: Formation and interpretation of the Bicoid morphogen gradient.Curr Top Dev Biol. 2020;137:79-117. doi: 10.1016/bs.ctdb.2019.11.016. Epub 2019 Dec 27. Curr Top Dev Biol. 2020. PMID: 32143754 Review.
-
Dynamic maternal gradients and morphogenetic networks in Drosophila early embryo.Biosystems. 2018 Nov;173:207-213. doi: 10.1016/j.biosystems.2018.10.009. Epub 2018 Oct 10. Biosystems. 2018. PMID: 30315821 Review.
Cited by
-
Modularity, criticality, and evolvability of a developmental gene regulatory network.Elife. 2019 Jun 6;8:e42832. doi: 10.7554/eLife.42832. Elife. 2019. PMID: 31169494 Free PMC article.
-
Speed and navigation control of thymocyte development by the fetal T-cell gene regulatory network.Immunol Rev. 2023 May;315(1):171-196. doi: 10.1111/imr.13190. Epub 2023 Feb 1. Immunol Rev. 2023. PMID: 36722494 Free PMC article. Review.
-
Speed regulation of genetic cascades allows for evolvability in the body plan specification of insects.Proc Natl Acad Sci U S A. 2017 Oct 10;114(41):E8646-E8655. doi: 10.1073/pnas.1702478114. Epub 2017 Sep 25. Proc Natl Acad Sci U S A. 2017. PMID: 28973882 Free PMC article.
-
Precision of tissue patterning is controlled by dynamical properties of gene regulatory networks.Development. 2021 Feb 25;148(4):dev197566. doi: 10.1242/dev.197566. Development. 2021. PMID: 33547135 Free PMC article.
-
Towards a physical understanding of developmental patterning.Nat Rev Genet. 2021 Aug;22(8):518-531. doi: 10.1038/s41576-021-00355-7. Epub 2021 May 10. Nat Rev Genet. 2021. PMID: 33972772 Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

