Evolution and Diversity of Transposable Elements in Vertebrate Genomes
- PMID: 28158585
- PMCID: PMC5381603
- DOI: 10.1093/gbe/evw264
Evolution and Diversity of Transposable Elements in Vertebrate Genomes
Abstract
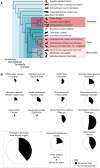
Transposable elements (TEs) are selfish genetic elements that mobilize in genomes via transposition or retrotransposition and often make up large fractions of vertebrate genomes. Here, we review the current understanding of vertebrate TE diversity and evolution in the context of recent advances in genome sequencing and assembly techniques. TEs make up 4-60% of assembled vertebrate genomes, and deeply branching lineages such as ray-finned fishes and amphibians generally exhibit a higher TE diversity than the more recent radiations of birds and mammals. Furthermore, the list of taxa with exceptional TE landscapes is growing. We emphasize that the current bottleneck in genome analyses lies in the proper annotation of TEs and provide examples where superficial analyses led to misleading conclusions about genome evolution. Finally, recent advances in long-read sequencing will soon permit access to TE-rich genomic regions that previously resisted assembly including the gigantic, TE-rich genomes of salamanders and lungfishes.
Keywords: retrotransposons; transposable element; transposons; vertebrate.
© The Author(s) 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Acosta MJ, Marchal JA, Fernández-Espartero CH, Bullejos M, Sánchez A. 2008. Retroelements (LINEs and SINEs) in vole genomes: differential distribution in the constitutive heterochromatin. Chromosome Res. 16:949–959. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

