An update on the Enzyme Portal: an integrative approach for exploring enzyme knowledge
- PMID: 28158609
- PMCID: PMC5421622
- DOI: 10.1093/protein/gzx008
An update on the Enzyme Portal: an integrative approach for exploring enzyme knowledge
Abstract
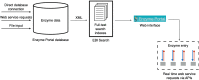
Enzymes are a key part of life processes and are increasingly important for various areas of research such as medicine, biotechnology, bioprocessing and drug research. The goal of the Enzyme Portal is to provide an interface to all European Bioinformatics Institute (EMBL-EBI) data about enzymes (de Matos, P., et al. , (2013), BMC Bioinformatics , (1), 103). These data include enzyme function, sequence features and family classification, protein structure, reactions, pathways, small molecules, diseases and the associated literature. The sources of enzyme data are: the UniProt Knowledgebase (UniProtKB) (UniProt Consortium, 2015), the Protein Data Bank in Europe (PDBe), (Valenkar, S., et al ., Nucleic Acids Res. 2016; , D385-D395) Rhea-a database of enzyme-catalysed reactions (Morgat, A., et al ., Nucleic Acids Res. 2015; , D459-D464), Reactome-a database of biochemical pathways (Fabregat, A., et al ., Nucleic Acids Res. 2016; , D481-D487), IntEnz-a resource with enzyme nomenclature information (Fleischmann, A., et al ., Nucleic Acids Res. 2004 , D434-D437) and ChEBI (Hastings, J., et al ., Nucleic Acids Res. 2013) and ChEMBL (Bento, A. P., et al ., Nucleic Acids Res. 2014 , 1083-1090)-resources which contain information about small-molecule chemistry and bioactivity. This article describes the redesign of Enzyme Portal and the increased functionality added to maximise integration and interpretation of these data. Use case examples of the Enzyme Portal and the versatile workflows its supports are illustrated. We welcome the suggestion of new resources for integration.
Keywords: Enzymes; integration; proteins; search; services.
© The Author 2017. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com
Figures





References
-
- Bento A.P., Gaulton A., Hersey A. et al. (2014) Nucleic Acids Res., 42, 1083–1090. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

