Abf1 and other general regulatory factors control ribosome biogenesis gene expression in budding yeast
- PMID: 28158860
- PMCID: PMC5416754
- DOI: 10.1093/nar/gkx058
Abf1 and other general regulatory factors control ribosome biogenesis gene expression in budding yeast
Abstract
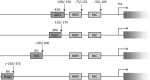
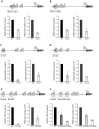
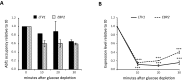
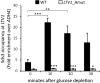
Ribosome biogenesis in Saccharomyces cerevisiae involves a regulon of >200 genes (Ribi genes) coordinately regulated in response to nutrient availability and cellular growth rate. Two cis-acting elements called PAC and RRPE are known to mediate Ribi gene repression in response to nutritional downshift. Here, we show that most Ribi gene promoters also contain binding sites for one or more General Regulatory Factors (GRFs), most frequently Abf1 and Reb1, and that these factors are enriched in vivo at Ribi promoters. Abf1/Reb1/Tbf1 promoter association was required for full Ribi gene expression in rich medium and for its modulation in response to glucose starvation, characterized by a rapid drop followed by slow recovery. Such a response did not entail changes in Abf1 occupancy, but it was paralleled by a quick increase, followed by slow decrease, in Rpd3L histone deacetylase occupancy. Remarkably, Abf1 site disruption also abolished Rpd3L complex recruitment in response to starvation. Extensive mutational analysis of the DBP7 promoter revealed a complex interplay of Tbf1 sites, PAC and RRPE in the transcriptional regulation of this Ribi gene. Our observations point to GRFs as new multifaceted players in Ribi gene regulation both during exponential growth and under repressive conditions.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Lempiainen H., Shore D.. Growth control and ribosome biogenesis. Curr. Opin. Cell Biol. 2009; 21:855–863. - PubMed
-
- Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999; 24:437–440. - PubMed
-
- Dieci G., Preti M., Montanini B.. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics. 2009; 94:83–88. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

