SETting the Stage for Cancer Development: SETD2 and the Consequences of Lost Methylation
- PMID: 28159833
- PMCID: PMC5411680
- DOI: 10.1101/cshperspect.a026468
SETting the Stage for Cancer Development: SETD2 and the Consequences of Lost Methylation
Abstract
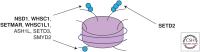
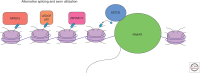
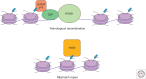
The H3 lysine 36 histone methyltransferase SETD2 is mutated across a range of human cancers. Although other enzymes can mediate mono- and dimethylation, SETD2 is the exclusive trimethylase. SETD2 associates with the phosphorylated carboxy-terminal domain of RNA polymerase and modifies histones at actively transcribed genes. The functions associated with SETD2 are mediated through multiple effector proteins that bind trimethylated H3K36. These effectors directly mediate multiple chromatin-regulated processes, including RNA splicing, DNA damage repair, and DNA methylation. Although alterations in each of these processes have been associated with SETD2 loss, the relative role of each in the development of cancer is not fully understood. Critical vulnerabilities resulting from SETD2 loss may offer a strategy for potential therapeutics.
Copyright © 2017 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures

References
-
- Bannister AJ, Schneider R, Myers F a, Thorne AW, Crane-Robinson C, Kouzarides T. 2005. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem 280: 17732–17736. - PubMed
-
- Baubec T, Colombo DF, Wirbelauer C, Schmidt J, Burger L, Krebs AR, Akalin A, Schübeler D. 2015. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 520: 243–247. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
