ATLAS: A database linking binding affinities with structures for wild-type and mutant TCR-pMHC complexes
- PMID: 28160322
- PMCID: PMC5860664
- DOI: 10.1002/prot.25260
ATLAS: A database linking binding affinities with structures for wild-type and mutant TCR-pMHC complexes
Abstract
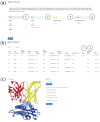
The ATLAS (Altered TCR Ligand Affinities and Structures) database (https://zlab.umassmed.edu/atlas/web/) is a manually curated repository containing the binding affinities for wild-type and mutant T cell receptors (TCRs) and their antigens, peptides presented by the major histocompatibility complex (pMHC). The database links experimentally measured binding affinities with the corresponding three dimensional (3D) structures for TCR-pMHC complexes. The user can browse and search affinities, structures, and experimental details for TCRs, peptides, and MHCs of interest. We expect this database to facilitate the development of next-generation protein design algorithms targeting TCR-pMHC interactions. ATLAS can be easily parsed using modeling software that builds protein structures for training and testing. As an example, we provide structural models for all mutant TCRs in ATLAS, built using the Rosetta program. Utilizing these structures, we report a correlation of 0.63 between experimentally measured changes in binding energies and our predicted changes. Proteins 2017; 85:908-916. © 2016 Wiley Periodicals, Inc.
Keywords: 3D viewer; Rosetta; adaptive immunity; binding energy; protein modeling; scoring functions.
© 2017 Wiley Periodicals, Inc.
Figures


References
-
- Zinkernagel RM, Doherty PC. Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature. 1974;248:701–710. - PubMed
-
- Babbitt BP, Allen PM, Matsueda G, Haber E, Unanue ER. Binding of immunogenic peptides to Ia histocompatibility molecules. Nature. 1985;317:359–361. - PubMed
-
- Varela-Rohena A, Molloy PE, Dunn SM, Li Y, Suhoski MM, Carroll RG, Milicic A, Mahon T, Sutton DH, Laugel B, Moysey R, Cameron BJ, Vuidepot A, Purbhoo MA, Cole DK, Phillips RE, June CH, Jakobsen BK, Sewell AK, Riley JL. Control of HIV-1 immune escape by CD8 T cells expressing enhanced T-cell receptor. Nat Med. 2008;14(12):1390–1395. - PMC - PubMed
-
- Linette GP, Stadtmauer Ea, Maus MV, Rapoport AP, Levine BL, Emery L, Litzky L, Bagg A, Carreno BM, Cimino PJ, Binder-Scholl GK, Smethurst DP, Gerry AB, Pumphrey NJ, Bennett AD, Brewer JE, Dukes J, Harper J, Tayton-Martin HK, Jakobsen BK, Hassan NJ, Kalos M, June CH. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood. 2013;122(6):863–871. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

