A novel PHD-finger protein 14/KIF4A complex overexpressed in lung cancer is involved in cell mitosis regulation and tumorigenesis
- PMID: 28160558
- PMCID: PMC5386714
- DOI: 10.18632/oncotarget.14962
A novel PHD-finger protein 14/KIF4A complex overexpressed in lung cancer is involved in cell mitosis regulation and tumorigenesis
Abstract
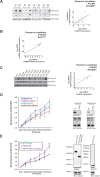
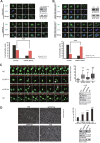
The plant homeodomain (PHD) finger-containing proteins have been implicated in many human diseases including cancer. In this study, we found that PHF14, a newly identified PHD finger protein, is highly expressed in lung cancer. The high expression level of PHF14 was associated with adenocarcinoma and poor survival in lung cancer patients. Knocking down PHF14 suppressed cancer cell growth and carcinogenesis, while over-expressing PHF14 promoted cell proliferation. During cell division, PHF14 directly bound to and co-localized with KIF4A (a nuclear motor protein involved in lung carcinogenesis) to form a functional complex. Similarly to the effect of KIF4A depletion, silencing PHF14 in several cell lines caused cell mitotic defects, prolonged M phase, and inhibited cell proliferation. What's more, these two proteins had a synergistic effect on cell proliferation and were significantly co-overexpressed in lung cancer tissues. Our data provide new insights into the biological significance of PHD finger proteins and imply that PHF14 may be a potential biomarker for lung cancer.
Keywords: KIF4A; PHF14; biomarker; lung carcinogenesis; mitosis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
TRIM47 overexpression is a poor prognostic factor and contributes to carcinogenesis in non-small cell lung carcinoma.Oncotarget. 2017 Apr 4;8(14):22730-22740. doi: 10.18632/oncotarget.15188. Oncotarget. 2017. PMID: 28186994 Free PMC article.
-
COPB2 promotes cell proliferation and tumorigenesis through up-regulating YAP1 expression in lung adenocarcinoma cells.Biomed Pharmacother. 2018 Jul;103:373-380. doi: 10.1016/j.biopha.2018.04.006. Epub 2018 Apr 24. Biomed Pharmacother. 2018. PMID: 29674272
-
Overexpression of KIF18A promotes cell proliferation, inhibits apoptosis, and independently predicts unfavorable prognosis in lung adenocarcinoma.IUBMB Life. 2019 Jul;71(7):942-955. doi: 10.1002/iub.2030. Epub 2019 Feb 28. IUBMB Life. 2019. PMID: 30817091
-
H2AX a promising biomarker for lung cancer: a review.Cancer Invest. 2013 Nov;31(9):582-99. doi: 10.3109/07357907.2013.849721. Cancer Invest. 2013. PMID: 24164298 Review.
-
Mouse lung epithelial cell lines--tools for the study of differentiation and the neoplastic phenotype.Toxicology. 1997 Nov 21;123(1-2):53-100. doi: 10.1016/s0300-483x(97)00108-x. Toxicology. 1997. PMID: 9347924 Review.
Cited by
-
Regulation of the mitotic chromosome folding machines.Biochem J. 2022 Oct 28;479(20):2153-2173. doi: 10.1042/BCJ20210140. Biochem J. 2022. PMID: 36268993 Free PMC article.
-
Impact of KIF4A on Cancer Stem Cells and EMT in Lung Cancer and Glioma.Cancers (Basel). 2023 Nov 22;15(23):5523. doi: 10.3390/cancers15235523. Cancers (Basel). 2023. PMID: 38067227 Free PMC article.
-
KIF4A Regulates the Progression of Pancreatic Ductal Adenocarcinoma through Proliferation and Invasion.Biomed Res Int. 2021 Nov 11;2021:8249293. doi: 10.1155/2021/8249293. eCollection 2021. Biomed Res Int. 2021. Retraction in: Biomed Res Int. 2025 Apr 29;2025:9759616. doi: 10.1155/bmri/9759616. PMID: 34805404 Free PMC article. Retracted.
-
Whole Exome Sequencing Identifies PHF14 Mutations in Neurocytoma and Predicts Responsivity to the PDGFR Inhibitor Sunitinib.Biomedicines. 2022 Nov 8;10(11):2842. doi: 10.3390/biomedicines10112842. Biomedicines. 2022. PMID: 36359362 Free PMC article.
-
Molecular basis for bipartite recognition of histone H3 by the PZP domain of PHF14.Nucleic Acids Res. 2021 Sep 7;49(15):8961-8973. doi: 10.1093/nar/gkab670. Nucleic Acids Res. 2021. PMID: 34365506 Free PMC article.
References
-
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. - PubMed
-
- Micci F, Gorunova L, Gatius S, Matias-Guiu X, Davidson B, Heim S, Panagopoulos L. MEAF6/PHF1 is a recurrent gene fusion in endometrial stromal sarcoma. Cancer Lett. 2014;347:75–78. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

