Arm-specific cleavage and mutation during reverse transcription of 2΄,5΄-branched RNA by Moloney murine leukemia virus reverse transcriptase
- PMID: 28160599
- PMCID: PMC5399748
- DOI: 10.1093/nar/gkx073
Arm-specific cleavage and mutation during reverse transcription of 2΄,5΄-branched RNA by Moloney murine leukemia virus reverse transcriptase
Abstract
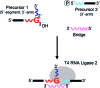
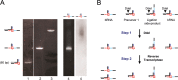
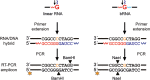
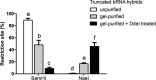
Branchpoint nucleotides of intron lariats induce pausing of DNA synthesis by reverse transcriptases (RTs), but it is not known yet how they direct RT RNase H activity on branched RNA (bRNA). Here, we report the effects of the two arms of bRNA on branchpoint-directed RNA cleavage and mutation produced by Moloney murine leukemia virus (M-MLV) RT during DNA polymerization. We constructed a long-chained bRNA template by splinted-ligation. The bRNA oligonucleotide is chimeric and contains DNA to identify RNA cleavage products by probe hybridization. Unique sequences surrounding the branchpoint facilitate monitoring of bRNA purification by terminal-restriction fragment length polymorphism analysis. We evaluate the M-MLV RT-generated cleavage and mutational patterns. We find that cleavage of bRNA and misprocessing of the branched nucleotide proceed arm-specifically. Bypass of the branchpoint from the 2΄-arm causes single-mismatch errors, whereas bypass from the 3΄-arm leads to deletion mutations. The non-template arm is cleaved when reverse transcription is primed from the 3΄-arm but not from the 2΄-arm. This suggests that RTs flip ∼180° at branchpoints and RNases H cleave the non-template arm depending on its accessibility. Our observed interplay between M-MLV RT and bRNA would be compatible with a bRNA-mediated control of retroviral and related retrotransposon replication.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



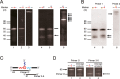

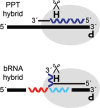
Similar articles
-
Dual coding potential of a 2',5'-branched ribonucleotide in DNA.RNA. 2019 Jan;25(1):105-120. doi: 10.1261/rna.068486.118. Epub 2018 Oct 25. RNA. 2019. PMID: 30361268 Free PMC article.
-
Similarities and differences in the RNase H activities of human immunodeficiency virus type 1 reverse transcriptase and Moloney murine leukemia virus reverse transcriptase.J Mol Biol. 1999 Dec 17;294(5):1097-113. doi: 10.1006/jmbi.1999.3325. J Mol Biol. 1999. PMID: 10600369
-
Mutations of the RNase H C helix of the Moloney murine leukemia virus reverse transcriptase reveal defects in polypurine tract recognition.J Virol. 2002 Aug;76(16):8360-73. doi: 10.1128/jvi.76.16.8360-8373.2002. J Virol. 2002. PMID: 12134040 Free PMC article.
-
RNase H activity: structure, specificity, and function in reverse transcription.Virus Res. 2008 Jun;134(1-2):86-103. doi: 10.1016/j.virusres.2007.12.007. Epub 2008 Feb 7. Virus Res. 2008. PMID: 18261820 Free PMC article. Review.
-
Murine leukemia virus reverse transcriptase: structural comparison with HIV-1 reverse transcriptase.Virus Res. 2008 Jun;134(1-2):186-202. doi: 10.1016/j.virusres.2008.01.001. Epub 2008 Feb 21. Virus Res. 2008. PMID: 18294720 Free PMC article. Review.
Cited by
-
Dual coding potential of a 2',5'-branched ribonucleotide in DNA.RNA. 2019 Jan;25(1):105-120. doi: 10.1261/rna.068486.118. Epub 2018 Oct 25. RNA. 2019. PMID: 30361268 Free PMC article.
-
Spontaneous advent of genetic diversity in RNA populations through multiple recombination mechanisms.RNA. 2019 Apr;25(4):453-464. doi: 10.1261/rna.068908.118. Epub 2019 Jan 22. RNA. 2019. PMID: 30670484 Free PMC article.
-
Saccharomyces cerevisiae RNA lariat debranching enzyme, Dbr1p, is required for completion of reverse transcription by the retrovirus-like element Ty1 and cleaves branched Ty1 RNAs.Mol Genet Genomics. 2021 Mar;296(2):409-422. doi: 10.1007/s00438-020-01753-y. Epub 2021 Jan 19. Mol Genet Genomics. 2021. PMID: 33464395
-
DGR mutagenic transposition occurs via hypermutagenic reverse transcription primed by nicked template RNA.Proc Natl Acad Sci U S A. 2017 Nov 21;114(47):E10187-E10195. doi: 10.1073/pnas.1715952114. Epub 2017 Nov 6. Proc Natl Acad Sci U S A. 2017. PMID: 29109248 Free PMC article.
References
-
- Le Grice S.F.J., Nowotny M.. Nucleic Acid Polymerases. 2014; 30:Berlin Heidelberg, NY: Springer-Verlag; 189–214.
-
- DeStefano J.J., Buiser R.G., Mallaber L.M., Bambara R.A., Fay P.J.. Human immunodeficiency virus reverse transcriptase displays a partially processive 3΄ to 5΄ endonuclease activity. J. Biol. Chem. 1991; 266:24295–24301. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

