eMouseAtlas: An atlas-based resource for understanding mammalian embryogenesis
- PMID: 28161522
- PMCID: PMC5442644
- DOI: 10.1016/j.ydbio.2017.01.023
eMouseAtlas: An atlas-based resource for understanding mammalian embryogenesis
Abstract
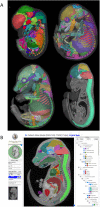
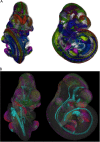
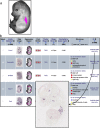
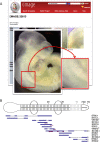
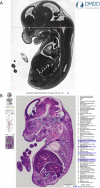
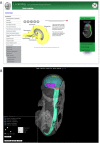
The eMouseAtlas resource is an online database of 3D digital models of mouse development, an ontology of mouse embryo anatomy and a gene-expression database with about 30K spatially mapped gene-expression patterns. It is closely linked with the MGI/GXD database at the Jackson Laboratory and holds links to almost all available image-based gene-expression data for the mouse embryo. In this resource article we describe the novel web-based tools we have developed for 3D visualisation of embryo anatomy and gene expression. We show how mapping of gene expression data onto spatial models delivers a framework for capturing gene expression that enhances our understanding of development, and we review the exploratory tools utilised by the EMAGE gene expression database as a means of defining co-expression of in situ hybridisation, immunohistochemistry, and lacZ-omic expression patterns. We report on recent developments of the eHistology atlas and our use of web-services to support embedding of the online 'The Atlas of Mouse Development' in the context of other resources such as the DMDD mouse phenotype database. In addition, we discuss new developments including a cellular-resolution placental atlas, third-party atlas models, clonal analysis data and a new interactive eLearning resource for developmental processes.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Attwood T.K., Agit B., Ellis L.B.M. Longevity of biological databases. EMBnet J. 2015
-
- Baldock R., Bard J., Davidson D.R., Morriss-Kay G. Elsevier; 2015. Kaufman's Atlas of Mouse Development.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

