Picornaviral polymerase structure, function, and fidelity modulation
- PMID: 28163093
- PMCID: PMC5476519
- DOI: 10.1016/j.virusres.2017.01.026
Picornaviral polymerase structure, function, and fidelity modulation
Abstract
Like all positive strand RNA viruses, the picornaviruses replicate their genomes using a virally encoded RNA-dependent RNA polymerase enzyme known as 3Dpol. Over the past decade we have made tremendous advances in our understanding of 3Dpol structure and function, including the discovery of a novel mechanism for closing the active site that allows these viruses to easily fine tune replication fidelity and quasispecies distributions. This review summarizes current knowledge of picornaviral polymerase structure and how the enzyme interacts with RNA and other viral proteins to form stable and processive elongation complexes. The picornaviral RdRPs are among the smallest viral polymerases, but their fundamental molecular mechanism for catalysis appears to be generally applicable as a common feature of all positive strand RNA virus polymerases.
Keywords: Picornavirus; Polymerase; Positive strand RNA virus; RNA-dependent RNA polymerase; Structure.
Copyright © 2017 Elsevier B.V. All rights reserved.
Figures


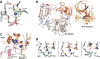


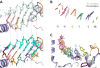

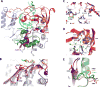
References
-
- Arias A, Arnold JJ, Sierra M, Smidansky ED, Domingo E, Cameron CE. Determinants of RNA-dependent RNA polymerase (in)fidelity revealed by kinetic analysis of the polymerase encoded by a foot-and-mouth disease virus mutant with reduced sensitivity to ribavirin. J Virol. 2008;82:12346–12355. - PMC - PubMed
-
- Arnold JJ, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3D(pol)). Assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub) J Biol Chem. 2000;275:5329–5336. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

