Use of application containers and workflows for genomic data analysis
- PMID: 28163975
- PMCID: PMC5248400
- DOI: 10.4103/2153-3539.197197
Use of application containers and workflows for genomic data analysis
Abstract
Background: The rapid acquisition of biological data and development of computationally intensive analyses has led to a need for novel approaches to software deployment. In particular, the complexity of common analytic tools for genomics makes them difficult to deploy and decreases the reproducibility of computational experiments.
Methods: Recent technologies that allow for application virtualization, such as Docker, allow developers and bioinformaticians to isolate these applications and deploy secure, scalable platforms that have the potential to dramatically increase the efficiency of big data processing.
Results: While limitations exist, this study demonstrates a successful implementation of a pipeline with several discrete software applications for the analysis of next-generation sequencing (NGS) data.
Conclusions: With this approach, we significantly reduced the amount of time needed to perform clonal analysis from NGS data in acute myeloid leukemia.
Keywords: Big data; bioinformatics workflow; containerization; genomics.
Conflict of interest statement
There are no conflicts of interest.
Figures
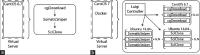
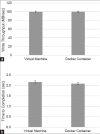

Similar articles
-
Bio-Docklets: virtualization containers for single-step execution of NGS pipelines.Gigascience. 2017 Aug 1;6(8):1-7. doi: 10.1093/gigascience/gix048. Gigascience. 2017. PMID: 28854616 Free PMC article.
-
ILIAD: a suite of automated Snakemake workflows for processing genomic data for downstream applications.BMC Bioinformatics. 2023 Nov 8;24(1):424. doi: 10.1186/s12859-023-05548-x. BMC Bioinformatics. 2023. PMID: 37940870 Free PMC article.
-
Scalable Workflows and Reproducible Data Analysis for Genomics.Methods Mol Biol. 2019;1910:723-745. doi: 10.1007/978-1-4939-9074-0_24. Methods Mol Biol. 2019. PMID: 31278683 Free PMC article.
-
Principles and Validation of Bioinformatics Pipeline for Cancer Next-Generation Sequencing.Clin Lab Med. 2022 Sep;42(3):409-421. doi: 10.1016/j.cll.2022.05.006. Epub 2022 Aug 22. Clin Lab Med. 2022. PMID: 36150820 Review.
-
Using R and Bioconductor in Clinical Genomics and Transcriptomics.J Mol Diagn. 2020 Jan;22(1):3-20. doi: 10.1016/j.jmoldx.2019.08.006. Epub 2019 Oct 9. J Mol Diagn. 2020. PMID: 31605800 Review.
Cited by
-
Hot-starting software containers for STAR aligner.Gigascience. 2018 Aug 1;7(8):giy092. doi: 10.1093/gigascience/giy092. Gigascience. 2018. PMID: 30085034 Free PMC article.
-
Reproducible Bioconductor workflows using browser-based interactive notebooks and containers.J Am Med Inform Assoc. 2018 Jan 1;25(1):4-12. doi: 10.1093/jamia/ocx120. J Am Med Inform Assoc. 2018. PMID: 29092073 Free PMC article.
-
PGSXplorer: an integrated nextflow pipeline for comprehensive quality control and polygenic score model development.PeerJ. 2025 Feb 12;13:e18973. doi: 10.7717/peerj.18973. eCollection 2025. PeerJ. 2025. PMID: 39959831 Free PMC article.
-
DockerBIO: web application for efficient use of bioinformatics Docker images.PeerJ. 2018 Nov 27;6:e5954. doi: 10.7717/peerj.5954. eCollection 2018. PeerJ. 2018. PMID: 30515360 Free PMC article.
-
A complete pedigree-based graph workflow for rare candidate variant analysis.Genome Res. 2022 May;32(5):893-903. doi: 10.1101/gr.276387.121. Epub 2022 Apr 28. Genome Res. 2022. PMID: 35483961 Free PMC article.
References
-
- Krumholz HM, Waldstreicher J. The Yale Open Data Access (YODA) Project – A mechanism for data sharing. N Engl J Med. 2016;375:403–5. - PubMed
-
- Collins FS, Barker AD. Mapping the cancer genome. Pinpointing the genes involved in cancer will help chart a new course across the complex landscape of human malignancies. Sci Am. 2007;296:50–7. - PubMed
-
- Nekrutenko A, Taylor J. Next-generation sequencing data interpretation: Enhancing reproducibility and accessibility. Nat Rev Genet. 2012;13:667–72. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

