Genome-wide data from two early Neolithic East Asian individuals dating to 7700 years ago
- PMID: 28164156
- PMCID: PMC5287702
- DOI: 10.1126/sciadv.1601877
Genome-wide data from two early Neolithic East Asian individuals dating to 7700 years ago
Abstract
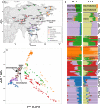
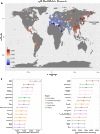
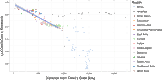
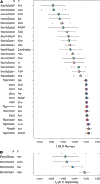
Ancient genomes have revolutionized our understanding of Holocene prehistory and, particularly, the Neolithic transition in western Eurasia. In contrast, East Asia has so far received little attention, despite representing a core region at which the Neolithic transition took place independently ~3 millennia after its onset in the Near East. We report genome-wide data from two hunter-gatherers from Devil's Gate, an early Neolithic cave site (dated to ~7.7 thousand years ago) located in East Asia, on the border between Russia and Korea. Both of these individuals are genetically most similar to geographically close modern populations from the Amur Basin, all speaking Tungusic languages, and, in particular, to the Ulchi. The similarity to nearby modern populations and the low levels of additional genetic material in the Ulchi imply a high level of genetic continuity in this region during the Holocene, a pattern that markedly contrasts with that reported for Europe.
Keywords: East Asia; Russian Far East; ancient genetics; human population genetics; neolithic.
Figures
References
-
- Lazaridis I., Nadel D., Rollefson G., Merrett D. C., Rohland N., Mallick S., Fernandes D., Novak M., Gamarra B., Sirak K., Connell S., Stewardson K., Harney E., Fu Q., Gonzalez-Fortes G., Jones E. R., Alpaslan Roodenberg S., Lengyel G., Bocquentin F., Gasparian B., Monge J. M., Gregg M., Eshed V., Mizrahi A.-S., Meiklejohn C., Gerritsen F., Bejenaru L., Blüher M., Campbell A., Cavalleri G., Comas D., Froguel P., Gilbert E., Kerr S. M., Kovacs P., Krause J., McGettigan D., Merrigan M., Andrew Merriwether D., O’Reilly S., Richards M. B., Semino O., Shamoon-Pour M., Stefanescu G., Stumvoll M., Tönjes A., Torroni A., Wilson J. F., Yengo L., Hovhannisyan N. A., Patterson N., Pinhasi R., Reich D., Genomic insights into the origin of farming in the ancient Near East. Nature 536, 419–424 (2016). - PMC - PubMed
-
- Gallego-Llorente M., Connell S., Jones E. R., Merrett D. C., Jeon Y., Eriksson A., Siska V., Gamba C., Meiklejohn C., Beyer R., Jeon S., Cho Y. S., Hofreiter M., Bhak J., Manica A., Pinhasi R., The genetics of an early Neolithic pastoralist from the Zagros, Iran. Sci. Rep. 6, 31326 (2016). - PMC - PubMed
-
- Lazaridis I., Patterson N., Mittnik A., Renaud G., Mallick S., Kirsanow K., Sudmant P. H., Schraiber J. G., Castellano S., Lipson M., Berger B., Economou C., Bollongino R., Fu Q., Bos K. I., Nordenfelt S., Li H., de Filippo C., Prüfer K., Sawyer S., Posth C., Haak W., Hallgren F., Fornander E., Rohland N., Delsate D., Francken M., Guinet J.-M., Wahl J., Ayodo G., Babiker H. A., Bailliet G., Balanovska E., Balanovsky O., Barrantes R., Bedoya G., Ben-Ami H., Bene J., Berrada F., Bravi C. M., Brisighelli F., Busby G. B. J., Cali F., Churnosov M., Cole D. E. C., Corach D., Damba L., van Driem G., Dryomov S., Dugoujon J.-M., Fedorova S. A., Gallego Romero I., Gubina M., Hammer M., Henn B. M., Hervig T., Hodoglugil U., Jha A. R., Karachanak-Yankova S., Khusainova R., Khusnutdinova E., Kittles R., Kivisild T., Klitz W., Kučinskas V., Kushniarevich A., Laredj L., Litvinov S., Loukidis T., Mahley R. W., Melegh B., Metspalu E., Molina J., Mountain J., Näkkäläjärvi K., Nesheva D., Nyambo T., Osipova L., Parik J., Platonov F., Posukh O., Romano V., Rothhammer F., Rudan I., Ruizbakiev R., Sahakyan H., Sajantila A., Salas A., Starikovskaya E. B., Tarekegn A., Toncheva D., Turdikulova S., Uktveryte I., Utevska O., Vasquez R., Villena M., Voevoda M., Winkler C. A., Yepiskoposyan L., Zalloua P., Zemunik T., Cooper A., Capelli C., Thomas M. G., Ruiz-Linares A., Tishkoff S. A., Singh L., Thangaraj K., Villems R., Comas D., Sukernik R., Metspalu M., Meyer M., Eichler E. E., Burger J., Slatkin M., Pääbo S., Kelso J., Reich D., Krause J., Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014). - PMC - PubMed
-
- Haak W., Lazaridis I., Patterson N., Rohland N., Mallick S., Llamas B., Brandt G., Nordenfelt S., Harney E., Stewardson K., Fu Q., Mittnik A., Bánffy E., Economou C., Francken M., Friederich S., Garrido Pena R., Hallgren F., Khartanovich V., Khokhlov A., Kunst M., Kuznetsov P., Meller H., Mochalov O., Moiseyev V., Nicklisch N., Pichler S. L., Risch R., Rojo Guerra M. A., Roth C., Szécsényi-Nagy A., Wahl J., Meyer M., Krause J., Brown D., Anthony D., Cooper A., Werner Alt K., Reich D., Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211 (2015). - PMC - PubMed
-
- Jones E. R., Gonzalez-Fortes G., Connell S., Siska V., Eriksson A., Martiniano R., McLaughlin R. L., Gallego Llorente M., Cassidy L. M., Gamba C., Meshveliani T., Bar-Yosef O., Müller W., Belfer-Cohen A., Matskevich Z., Jakeli N., Higham T. F. G., Currat M., Lordkipanidze D., Hofreiter M., Manica A., Pinhasi R., Bradley D. G., Upper Palaeolithic genomes reveal deep roots of modern Eurasians. Nat. Commun. 6, 8912 (2015). - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

