Developmentally regulated alternative splicing is perturbed in type 1 diabetic skeletal muscle
- PMID: 28164326
- PMCID: PMC5704903
- DOI: 10.1002/mus.25599
Developmentally regulated alternative splicing is perturbed in type 1 diabetic skeletal muscle
Abstract
Introduction: Type 1 diabetic patients can develop skeletal muscle weakness and atrophy by molecular mechanisms that are not well understood. Alternative splicing (AS) is critical for gene expression in the skeletal muscle, and its dysregulation is implicated in muscle weakness and atrophy. Therefore, we investigated whether AS patterns are affected in type 1 diabetic skeletal muscle contributing to skeletal muscle defects.
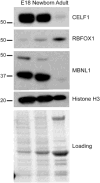
Methods: AS patterns were determined by reverse transcription-polymerase chain reaction and levels of RNA binding proteins were assessed by Western blot in type 1 diabetic mouse skeletal muscle and during normal mouse skeletal muscle development.
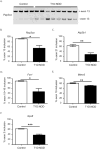
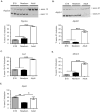
Results: Five genes with critical functions in the skeletal muscle are misspliced in type 1 diabetic skeletal muscle, resembling their AS patterns at embryonic stages. AS of these genes undergoes dramatic transitions during skeletal muscle development, correlating with changes in specific RNA binding proteins.
Conclusion: Embryonic spliced variants are inappropriately expressed in type 1 diabetic skeletal muscle. Muscle Nerve 56: 744-749, 2017.
Keywords: RNA binding proteins; alternative splicing; muscle development; skeletal muscle; type 1 diabetes.
© 2017 Wiley Periodicals, Inc.
Figures

Comment in
-
Reversion to embryonic transcriptional splicing patterns may underlie diabetic myopathy.Muscle Nerve. 2017 Oct;56(4):686-688. doi: 10.1002/mus.25745. Epub 2017 Aug 29. Muscle Nerve. 2017. PMID: 28771754 No abstract available.
References
-
- Aiello LP, Gardner TW, King GL, Blankenship G, Cavallerano JD, Ferris FL, 3rd, et al. Diabetic retinopathy. Diabetes Care. 1998;21:143–156. - PubMed
-
- Huynh K, Bernardo BC, McMullen JR, Ritchie RH. Diabetic cardiomyopathy: mechanisms and new treatment strategies targeting antioxidant signaling pathways. Pharmacol Ther. 2014;142:375–415. - PubMed
-
- Coppini DV. Enigma of painful diabetic neuropathy: can we use the basic science, research outcomes and real-world data to help improve patient care and outcomes? Diabet Med. 2016;33:1477–1482. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

