Application of circulating tumor DNA in prospective clinical oncology trials - standardization of preanalytical conditions
- PMID: 28164427
- PMCID: PMC5527445
- DOI: 10.1002/1878-0261.12037
Application of circulating tumor DNA in prospective clinical oncology trials - standardization of preanalytical conditions
Abstract
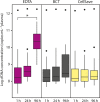
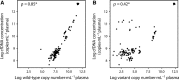
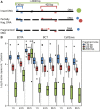
Circulating tumor DNA (ctDNA) has emerged as a potential new biomarker with diagnostic, predictive, and prognostic applications for various solid tumor types. Before beginning large prospective clinical trials to prove the added value of utilizing ctDNA in clinical practice, it is essential to investigate the effects of various preanalytical conditions on the quality of cell-free DNA (cfDNA) in general and of ctDNA in particular in order to optimize and standardize these conditions. Whole blood samples were collected from patients with metastatic cancer bearing a known somatic variant. The following preanalytical conditions were investigated: (a) different time intervals to plasma isolation (1, 24, and 96 h) and (b) different preservatives in blood collection tubes (EDTA, CellSave, and BCT). The quality of cfDNA/ctDNA was assessed by DNA quantification, digital polymerase chain reaction (dPCR) for somatic variant detection and a β-actin fragmentation assay for DNA contamination from lysed leukocytes. In 11 (69%) of our 16 patients, we were able to detect the known somatic variant in ctDNA. We observed a time-dependent increase in cfDNA concentrations in EDTA tubes, which was positively correlated with an increase in wild-type copy numbers and large DNA fragments (> 420 bp). Using different preservatives did not affect somatic variant detection ability, but did stabilize cfDNA concentrations over time. Variant allele frequency was affected by fluctuations in cfDNA concentration only in EDTA tubes at 96 h. Both CellSave and BCT tubes ensured optimal ctDNA quality in plasma processed within 96 h after blood collection for downstream somatic variant detection by dPCR.
Keywords: DNA contamination; blood collection tube; cell-free DNA; circulating tumor DNA; preanalytical condition.
© 2017 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Figures


Similar articles
-
Comparative analysis of circulating tumor DNA stability In K3EDTA, Streck, and CellSave blood collection tubes.Clin Biochem. 2016 Dec;49(18):1354-1360. doi: 10.1016/j.clinbiochem.2016.03.012. Epub 2016 Apr 27. Clin Biochem. 2016. PMID: 27129799
-
The Effect of Preservative and Temperature on the Analysis of Circulating Tumor DNA.Clin Cancer Res. 2017 May 15;23(10):2471-2477. doi: 10.1158/1078-0432.CCR-16-1691. Epub 2016 Nov 8. Clin Cancer Res. 2017. PMID: 27827317
-
A Study of Pre-Analytical Variables and Optimization of Extraction Method for Circulating Tumor DNA Measurements by Digital Droplet PCR.Cancer Epidemiol Biomarkers Prev. 2019 May;28(5):909-916. doi: 10.1158/1055-9965.EPI-18-0586. Epub 2019 Mar 1. Cancer Epidemiol Biomarkers Prev. 2019. PMID: 30824523
-
Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review.J Clin Oncol. 2018 Jun 1;36(16):1631-1641. doi: 10.1200/JCO.2017.76.8671. Epub 2018 Mar 5. J Clin Oncol. 2018. PMID: 29504847 Review.
-
Potential of modern circulating cell-free DNA diagnostic tools for detection of specific tumour cells in clinical practice.Biochem Med (Zagreb). 2020 Oct 15;30(3):030504. doi: 10.11613/BM.2020.030504. Epub 2020 Aug 5. Biochem Med (Zagreb). 2020. PMID: 32774122 Free PMC article. Review.
Cited by
-
Potential value of detection of minimal residual disease in colorectal cancer following radical resection.Chin J Cancer Res. 2024 Aug 30;36(4):442-454. doi: 10.21147/j.issn.1000-9604.2024.04.07. Chin J Cancer Res. 2024. PMID: 39246709 Free PMC article.
-
Detection of circulating tumour DNA after neoadjuvant chemoradiotherapy in patients with locally advanced oesophageal cancer.J Pathol. 2023 Jan;259(1):35-45. doi: 10.1002/path.6016. Epub 2022 Oct 31. J Pathol. 2023. PMID: 36196486 Free PMC article.
-
RAS and BRAF mutations in cell-free DNA are predictive for outcome of cetuximab monotherapy in patients with tissue-tested RAS wild-type advanced colorectal cancer.Mol Oncol. 2019 Nov;13(11):2361-2374. doi: 10.1002/1878-0261.12550. Epub 2019 Sep 30. Mol Oncol. 2019. PMID: 31350822 Free PMC article.
-
Cancer Diagnosis Using a Liquid Biopsy: Challenges and Expectations.Diagnostics (Basel). 2018 May 9;8(2):31. doi: 10.3390/diagnostics8020031. Diagnostics (Basel). 2018. PMID: 29747380 Free PMC article. Review.
-
Profiling (placental) DNA methylation in cell-free DNA across gestation: the Rotterdam Periconception Cohort.Mol Hum Reprod. 2025 Apr 3;31(2):gaaf011. doi: 10.1093/molehr/gaaf011. Mol Hum Reprod. 2025. PMID: 40338255 Free PMC article.
References
-
- Bidard FC, Madic J, Mariani P, Piperno‐Neumann S, Rampanou A, Servois V, Cassoux N, Desjardins L, Milder M, Vaucher I et al (2014) Detection rate and prognostic value of circulating tumor cells and circulating tumor DNA in metastatic uveal melanoma. Int J Cancer 134, 1207–1213. - PubMed
-
- Bortner CD, Oldenburg NB and Cidlowski JA (1995) The role of DNA fragmentation in apoptosis. Trends Cell Biol 5, 21–26. - PubMed
-
- Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler‐Araujo B et al (2013) Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 368, 1199–1209. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

