Structural analysis and evolution of specificity of the SUMO UFD E1-E2 interactions
- PMID: 28165030
- PMCID: PMC5292753
- DOI: 10.1038/srep41998
Structural analysis and evolution of specificity of the SUMO UFD E1-E2 interactions
Abstract
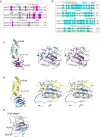
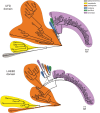
SUMO belongs to the ubiquitin-like family (UbL) of protein modifiers. SUMO is conserved among eukaryotes and is essential for the regulation of processes such as DNA damage repair, transcription, DNA replication and mitosis. UbL modification of proteins occurs via a specific enzymatic cascade formed by the crosstalk between the E1-activating enzyme, the E2-conjugating enzyme and the E3-ligase. An essential discrimination step in all UbL modifiers corresponds to the interaction between E1 and E2 enzymes, which is mediated by the recruitment of the E2 to the UFD domain (Ubiquitin-Fold Domain) of the E1 enzyme. To gain insights in the properties of this interface, we have compared the structures of the complexes between E1 UFD domain and E2 in human and yeast, revealing two alternative UFD platforms that interact with a conserved E2. Comparative sequence analysis of the E1 UFD domain indicates that the E2 binding region has been conserved across phylogenetic closely related species, in which higher sequence conservation can be found in the E2 binding region than in the entire UFD domain. These distinctive strategies for E1-E2 interactions through the UFD domain might be the consequence of a high selective pressure to ensure specificity of each modifier conjugation system.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Similar articles
-
Crystal structure of UBA2(ufd)-Ubc9: insights into E1-E2 interactions in Sumo pathways.PLoS One. 2010 Dec 30;5(12):e15805. doi: 10.1371/journal.pone.0015805. PLoS One. 2010. PMID: 21209884 Free PMC article.
-
Structural insights into SUMO E1-E2 interactions in Arabidopsis uncovers a distinctive platform for securing SUMO conjugation specificity across evolution.Biochem J. 2019 Jul 31;476(14):2127-2139. doi: 10.1042/BCJ20190232. Biochem J. 2019. PMID: 31292170
-
Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes.Cell. 2008 Jul 25;134(2):268-78. doi: 10.1016/j.cell.2008.05.046. Cell. 2008. PMID: 18662542
-
Protein interactions in the sumoylation cascade: lessons from X-ray structures.FEBS J. 2008 Jun;275(12):3003-15. doi: 10.1111/j.1742-4658.2008.06459.x. Epub 2008 May 17. FEBS J. 2008. PMID: 18492068 Review.
-
Molecular mechanisms in SUMO conjugation.Biochem Soc Trans. 2020 Feb 28;48(1):123-135. doi: 10.1042/BST20190357. Biochem Soc Trans. 2020. PMID: 31872228 Review.
Cited by
-
(De)Activation (Ir)Reversibly or Degradation: Dynamics of Post-Translational Protein Modifications in Plants.Life (Basel). 2022 Feb 21;12(2):324. doi: 10.3390/life12020324. Life (Basel). 2022. PMID: 35207610 Free PMC article. Review.
-
Insights into the transcriptional and post-transcriptional regulation of the rice SUMOylation machinery and into the role of two rice SUMO proteases.BMC Plant Biol. 2018 Dec 12;18(1):349. doi: 10.1186/s12870-018-1547-3. BMC Plant Biol. 2018. PMID: 30541427 Free PMC article.
-
SUMO chain formation relies on the amino-terminal region of SUMO-conjugating enzyme and has dedicated substrates in plants.Biochem J. 2018 Jan 2;475(1):61-74. doi: 10.1042/BCJ20170472. Biochem J. 2018. PMID: 29133528 Free PMC article.
-
Not So Slim Anymore-Evidence for the Role of SUMO in the Regulation of Lipid Metabolism.Biomolecules. 2020 Aug 6;10(8):1154. doi: 10.3390/biom10081154. Biomolecules. 2020. PMID: 32781719 Free PMC article. Review.
-
SUMOylation in Phytopathogen Interactions: Balancing Invasion and Resistance.Front Cell Dev Biol. 2021 Aug 16;9:703795. doi: 10.3389/fcell.2021.703795. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34485289 Free PMC article. Review.
References
-
- Johnson E. S. Protein modification by SUMO. Annual review of biochemistry 73, 355–382 (2004). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

