Architecture of the Vibrio cholerae toxin-coregulated pilus machine revealed by electron cryotomography
- PMID: 28165453
- PMCID: PMC5302817
- DOI: 10.1038/nmicrobiol.2016.269
Architecture of the Vibrio cholerae toxin-coregulated pilus machine revealed by electron cryotomography
Abstract
Type IV pili (T4P) are filamentous appendages found on many Bacteria and Archaea. They are helical fibres of pilin proteins assembled by a multi-component macromolecular machine we call the basal body. Based on pilin features, T4P are classified into type IVa pili (T4aP) and type IVb pili (T4bP)1,2. T4aP are more widespread and are involved in cell motility3, DNA transfer4, host predation5 and electron transfer6. T4bP are less prevalent and are mainly found in enteropathogenic bacteria, where they play key roles in host colonization7. Following similar work on T4aP machines8,9, here we use electron cryotomography10 to reveal the three-dimensional in situ structure of a T4bP machine in its piliated and non-piliated states. The specific machine we analyse is the Vibrio cholerae toxin-coregulated pilus machine (TCPM). Although only about half of the components of the TCPM show sequence homology to components of the previously analysed Myxococcus xanthus T4aP machine (T4aPM), we find that their structures are nevertheless remarkably similar. Based on homologies with components of the M. xanthus T4aPM and additional reconstructions of TCPM mutants in which the non-homologous proteins are individually deleted, we propose locations for all eight TCPM components within the complex. Non-homologous proteins in the T4aPM and TCPM are found to form similar structures, suggesting new hypotheses for their functions and evolutionary histories.
Figures


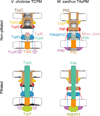
References
-
- Strom MS, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. - PubMed
-
- Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2:363–378. - PubMed
-
- Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. - PubMed
-
- Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat Rev Microbiol. 2004;2:241–249. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

