Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform
- PMID: 28165460
- PMCID: PMC5302332
- DOI: 10.1038/nmicrobiol.2016.274
Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform
Abstract
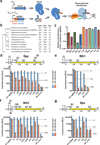
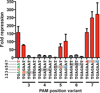
The development of new drug regimens that allow rapid, sterilizing treatment of tuberculosis has been limited by the complexity and time required for genetic manipulations in Mycobacterium tuberculosis. CRISPR interference (CRISPRi) promises to be a robust, easily engineered and scalable platform for regulated gene silencing. However, in M. tuberculosis, the existing Streptococcus pyogenes Cas9-based CRISPRi system is of limited utility because of relatively poor knockdown efficiency and proteotoxicity. To address these limitations, we screened eleven diverse Cas9 orthologues and identified four that are broadly functional for targeted gene knockdown in mycobacteria. The most efficacious of these proteins, the CRISPR1 Cas9 from Streptococcus thermophilus (dCas9Sth1), typically achieves 20- to 100-fold knockdown of endogenous gene expression with minimal proteotoxicity. In contrast to other CRISPRi systems, dCas9Sth1-mediated gene knockdown is robust when targeted far from the transcriptional start site, thereby allowing high-resolution dissection of gene function in the context of bacterial operons. We demonstrate the utility of this system by addressing persistent controversies regarding drug synergies in the mycobacterial folate biosynthesis pathway. We anticipate that the dCas9Sth1 CRISPRi system will have broad utility for functional genomics, genetic interaction mapping and drug-target profiling in M. tuberculosis.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

