STAT2 is an essential adaptor in USP18-mediated suppression of type I interferon signaling
- PMID: 28165510
- PMCID: PMC5365074
- DOI: 10.1038/nsmb.3378
STAT2 is an essential adaptor in USP18-mediated suppression of type I interferon signaling
Abstract
Type I interferons (IFNs) are multifunctional cytokines that regulate immune responses and cellular functions but also can have detrimental effects on human health. A tight regulatory network therefore controls IFN signaling, which in turn may interfere with medical interventions. The JAK-STAT signaling pathway transmits the IFN extracellular signal to the nucleus, thus resulting in alterations in gene expression. STAT2 is a well-known essential and specific positive effector of type I IFN signaling. Here, we report that STAT2 is also a previously unrecognized, crucial component of the USP18-mediated negative-feedback control in both human and mouse cells. We found that STAT2 recruits USP18 to the type I IFN receptor subunit IFNAR2 via its constitutive membrane-distal STAT2-binding site. This mechanistic coupling of effector and negative-feedback functions of STAT2 may provide novel strategies for treatment of IFN-signaling-related human diseases.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
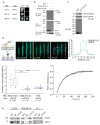

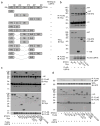
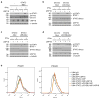
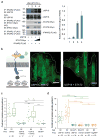
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

