Structural Analysis of Chemokine Receptor-Ligand Interactions
- PMID: 28165741
- PMCID: PMC5483895
- DOI: 10.1021/acs.jmedchem.6b01309
Structural Analysis of Chemokine Receptor-Ligand Interactions
Abstract
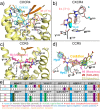
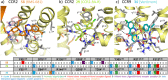
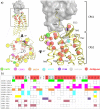
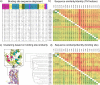
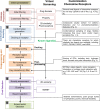
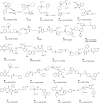
This review focuses on the construction and application of structural chemokine receptor models for the elucidation of molecular determinants of chemokine receptor modulation and the structure-based discovery and design of chemokine receptor ligands. A comparative analysis of ligand binding pockets in chemokine receptors is presented, including a detailed description of the CXCR4, CCR2, CCR5, CCR9, and US28 X-ray structures, and their implication for modeling molecular interactions of chemokine receptors with small-molecule ligands, peptide ligands, and large antibodies and chemokines. These studies demonstrate how the integration of new structural information on chemokine receptors with extensive structure-activity relationship and site-directed mutagenesis data facilitates the prediction of the structure of chemokine receptor-ligand complexes that have not been crystallized. Finally, a review of structure-based ligand discovery and design studies based on chemokine receptor crystal structures and homology models illustrates the possibilities and challenges to find novel ligands for chemokine receptors.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
















References
-
- Bongers G.; Maussang D.; Muniz L. R.; Noriega V. M.; Fraile-Ramos A.; Barker N.; Marchesi F.; Thirunarayanan N.; Vischer H. F.; Qin L.; Mayer L.; Harpaz N.; Leurs R.; Furtado G. C.; Clevers H.; Tortorella D.; Smit M. J.; Lira S. A. The cytomegalovirus-encoded chemokine receptor US28 promotes intestinal neoplasia in transgenic mice. J. Clin. Invest. 2010, 120, 3969–78. 10.1172/JCI42563. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information

