Divergent effects of intrinsically active MEK variants on developmental Ras signaling
- PMID: 28166211
- PMCID: PMC5621734
- DOI: 10.1038/ng.3780
Divergent effects of intrinsically active MEK variants on developmental Ras signaling
Abstract
Germline mutations in Ras pathway components are associated with a large class of human developmental abnormalities, known as RASopathies, that are characterized by a range of structural and functional phenotypes, including cardiac defects and neurocognitive delays. Although it is generally believed that RASopathies are caused by altered levels of pathway activation, the signaling changes in developing tissues remain largely unknown. We used assays with spatiotemporal resolution in Drosophila melanogaster (fruit fly) and Danio rerio (zebrafish) to quantify signaling changes caused by mutations in MAP2K1 (encoding MEK), a core component of the Ras pathway that is mutated in both RASopathies and cancers in humans. Surprisingly, we discovered that intrinsically active MEK variants can both increase and reduce the levels of pathway activation in vivo. The sign of the effect depends on cellular context, implying that some of the emerging phenotypes in RASopathies may be caused by increased, as well as attenuated, levels of Ras signaling.
Figures
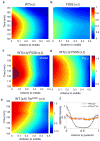
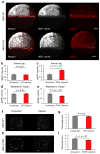
Similar articles
-
The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation.Curr Opin Genet Dev. 2009 Jun;19(3):230-6. doi: 10.1016/j.gde.2009.04.001. Epub 2009 May 19. Curr Opin Genet Dev. 2009. PMID: 19467855 Free PMC article. Review.
-
RASopathies: From germline mutations to somatic and multigenic diseases.Biomed J. 2021 Aug;44(4):422-432. doi: 10.1016/j.bj.2021.06.004. Epub 2021 Jun 24. Biomed J. 2021. PMID: 34175492 Free PMC article. Review.
-
In vivo severity ranking of Ras pathway mutations associated with developmental disorders.Proc Natl Acad Sci U S A. 2017 Jan 17;114(3):510-515. doi: 10.1073/pnas.1615651114. Epub 2017 Jan 3. Proc Natl Acad Sci U S A. 2017. PMID: 28049852 Free PMC article.
-
The RASopathies as an example of RAS/MAPK pathway disturbances - clinical presentation and molecular pathogenesis of selected syndromes.Dev Period Med. 2014 Jul-Sep;18(3):285-96. Dev Period Med. 2014. PMID: 25182392 Review.
-
Mosaic RASopathies.Cell Cycle. 2013 Jan 1;12(1):43-50. doi: 10.4161/cc.23108. Epub 2012 Dec 19. Cell Cycle. 2013. PMID: 23255105 Free PMC article. Review.
Cited by
-
Outstanding questions in developmental ERK signaling.Development. 2018 Jul 26;145(14):dev143818. doi: 10.1242/dev.143818. Development. 2018. PMID: 30049820 Free PMC article. Review.
-
Preformation and epigenesis converge to specify primordial germ cell fate in the early Drosophila embryo.PLoS Genet. 2022 Jan 5;18(1):e1010002. doi: 10.1371/journal.pgen.1010002. eCollection 2022 Jan. PLoS Genet. 2022. PMID: 34986144 Free PMC article.
-
Patient-derived iPSCs show premature neural differentiation and neuron type-specific phenotypes relevant to neurodevelopment.Mol Psychiatry. 2018 Aug;23(8):1687-1698. doi: 10.1038/mp.2017.238. Epub 2017 Nov 21. Mol Psychiatry. 2018. PMID: 29158583 Free PMC article.
-
How activating mutations affect MEK1 regulation and function.J Biol Chem. 2017 Nov 17;292(46):18814-18820. doi: 10.1074/jbc.C117.806067. Epub 2017 Oct 10. J Biol Chem. 2017. PMID: 29018093 Free PMC article.
-
Molecular mechanisms underlying cellular effects of human MEK1 mutations.Mol Biol Cell. 2021 Apr 19;32(9):974-983. doi: 10.1091/mbc.E20-10-0625. Epub 2021 Jan 21. Mol Biol Cell. 2021. PMID: 33476180 Free PMC article.
References
-
- Tartaglia M, et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001;29:465–468. - PubMed
-
- Rodriguez-Viciana P, et al. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science. 2006;311:1287–90. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

