Activated Oncogenic Pathway Modifies Iron Network in Breast Epithelial Cells: A Dynamic Modeling Perspective
- PMID: 28166223
- PMCID: PMC5293201
- DOI: 10.1371/journal.pcbi.1005352
Activated Oncogenic Pathway Modifies Iron Network in Breast Epithelial Cells: A Dynamic Modeling Perspective
Abstract
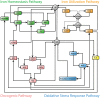
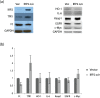
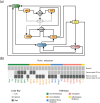
Dysregulation of iron metabolism in cancer is well documented and it has been suggested that there is interdependence between excess iron and increased cancer incidence and progression. In an effort to better understand the linkages between iron metabolism and breast cancer, a predictive mathematical model of an expanded iron homeostasis pathway was constructed that includes species involved in iron utilization, oxidative stress response and oncogenic pathways. The model leads to three predictions. The first is that overexpression of iron regulatory protein 2 (IRP2) recapitulates many aspects of the alterations in free iron and iron-related proteins in cancer cells without affecting the oxidative stress response or the oncogenic pathways included in the model. This prediction was validated by experimentation. The second prediction is that iron-related proteins are dramatically affected by mitochondrial ferritin overexpression. This prediction was validated by results in the pertinent literature not used for model construction. The third prediction is that oncogenic Ras pathways contribute to altered iron homeostasis in cancer cells. This prediction was validated by a combination of simulation experiments of Ras overexpression and catalase knockout in conjunction with the literature. The model successfully captures key aspects of iron metabolism in breast cancer cells and provides a framework upon which more detailed models can be built.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Knock-in of mutant K-ras in nontumorigenic human epithelial cells as a new model for studying K-ras mediated transformation.Cancer Res. 2007 Sep 15;67(18):8460-7. doi: 10.1158/0008-5472.CAN-07-0108. Cancer Res. 2007. PMID: 17875684
-
Oncogenic RAS alters the global and gene-specific histone modification pattern during epithelial-mesenchymal transition in colorectal carcinoma cells.Int J Biochem Cell Biol. 2010 Jun;42(6):911-20. doi: 10.1016/j.biocel.2010.01.024. Epub 2010 Jan 28. Int J Biochem Cell Biol. 2010. PMID: 20109579
-
ISGylation governs the oncogenic function of Ki-Ras in breast cancer.Oncogene. 2014 Feb 6;33(6):794-803. doi: 10.1038/onc.2012.633. Epub 2013 Jan 14. Oncogene. 2014. PMID: 23318454
-
Iron regulatory proteins, iron responsive elements and iron homeostasis.J Nutr. 1998 Dec;128(12):2295-8. doi: 10.1093/jn/128.12.2295. J Nutr. 1998. PMID: 9868172 Review.
-
Does excess iron play a role in breast carcinogenesis? An unresolved hypothesis.Cancer Causes Control. 2007 Dec;18(10):1047-53. doi: 10.1007/s10552-007-9058-9. Epub 2007 Sep 6. Cancer Causes Control. 2007. PMID: 17823849 Review.
Cited by
-
Control of Intracellular Molecular Networks Using Algebraic Methods.Bull Math Biol. 2019 Dec 23;82(1):2. doi: 10.1007/s11538-019-00679-w. Bull Math Biol. 2019. PMID: 31919596 Free PMC article.
-
Doxorubicin-transferrin conjugate alters mitochondrial homeostasis and energy metabolism in human breast cancer cells.Sci Rep. 2021 Feb 25;11(1):4544. doi: 10.1038/s41598-021-84146-4. Sci Rep. 2021. PMID: 33633284 Free PMC article.
-
A kinetic model of iron trafficking in growing Saccharomyces cerevisiae cells; applying mathematical methods to minimize the problem of sparse data and generate viable autoregulatory mechanisms.PLoS Comput Biol. 2023 Dec 19;19(12):e1011701. doi: 10.1371/journal.pcbi.1011701. eCollection 2023 Dec. PLoS Comput Biol. 2023. PMID: 38113197 Free PMC article.
-
A mathematical model of iron import and trafficking in wild-type and Mrs3/4ΔΔ yeast cells.BMC Syst Biol. 2019 Feb 21;13(1):23. doi: 10.1186/s12918-019-0702-2. BMC Syst Biol. 2019. PMID: 30791941 Free PMC article.
-
Transferrin receptor in primary and metastatic breast cancer: Evaluation of expression and experimental modulation to improve molecular targeting.PLoS One. 2023 Dec 20;18(12):e0293700. doi: 10.1371/journal.pone.0293700. eCollection 2023. PLoS One. 2023. PMID: 38117806 Free PMC article.
References
-
- Pinnix ZK, Miller LD, Wang W, D’Agostino R, Kute T, Willingham MC, et al. Ferroportin and Iron Regulation in Breast Cancer Progression and Prognosis. Science translational medicine. 2010. 08;2(43):43ra56–43ra56. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3734848/. 10.1126/scisignal.3001127 - DOI - PMC - PubMed
-
- Chifman J, Kniss A, Neupane P, Williams I, Leung B, Deng Z, et al. The core control system of intracellular iron homeostasis: A mathematical model. Journal of Theoretical Biology. 2012;300(0):91–99. Available from: http://www.sciencedirect.com/science/article/pii/S0022519312000343. 10.1016/j.jtbi.2012.01.024 - DOI - PMC - PubMed
-
- Epsztejn S, Kakhlon O, Glickstein H, Breuer W, Cabantchik ZI. Fluorescence Analysis of the Labile Iron Pool of Mammalian Cells. Analytical Biochemistry. 1997;248(1):31–40. Available from: http://www.sciencedirect.com/science/article/pii/S0003269797921266. 10.1006/abio.1997.2126 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

