Improving protein-protein interaction prediction using evolutionary information from low-quality MSAs
- PMID: 28166227
- PMCID: PMC5293240
- DOI: 10.1371/journal.pone.0169356
Improving protein-protein interaction prediction using evolutionary information from low-quality MSAs
Abstract
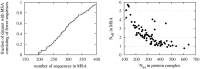
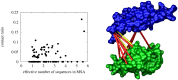
Evolutionary information stored in multiple sequence alignments (MSAs) has been used to identify the interaction interface of protein complexes, by measuring either co-conservation or co-mutation of amino acid residues across the interface. Recently, maximum entropy related correlated mutation measures (CMMs) such as direct information, decoupling direct from indirect interactions, have been developed to identify residue pairs interacting across the protein complex interface. These studies have focussed on carefully selected protein complexes with large, good-quality MSAs. In this work, we study protein complexes with a more typical MSA consisting of fewer than 400 sequences, using a set of 79 intramolecular protein complexes. Using a maximum entropy based CMM at the residue level, we develop an interface level CMM score to be used in re-ranking docking decoys. We demonstrate that our interface level CMM score compares favourably to the complementarity trace score, an evolutionary information-based score measuring co-conservation, when combined with the number of interface residues, a knowledge-based potential and the variability score of individual amino acid sites. We also demonstrate, that, since co-mutation and co-complementarity in the MSA contain orthogonal information, the best prediction performance using evolutionary information can be achieved by combining the co-mutation information of the CMM with co-conservation information of a complementarity trace score, predicting a near-native structure as the top prediction for 41% of the dataset. The method presented is not restricted to small MSAs, and will likely improve interface prediction also for complexes with large and good-quality MSAs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Predicting protein β-sheet contacts using a maximum entropy-based correlated mutation measure.Bioinformatics. 2013 Mar 1;29(5):580-7. doi: 10.1093/bioinformatics/btt005. Epub 2013 Jan 10. Bioinformatics. 2013. PMID: 23314126
-
Lessons from (co-)evolution in the docking of proteins and peptides for CAPRI Rounds 28-35.Proteins. 2017 Mar;85(3):378-390. doi: 10.1002/prot.25180. Epub 2016 Oct 24. Proteins. 2017. PMID: 27701780
-
Residue conservation information for generating near-native structures in protein-protein docking.J Bioinform Comput Biol. 2006 Aug;4(4):793-806. doi: 10.1142/s0219720006002223. J Bioinform Comput Biol. 2006. PMID: 17007068
-
A decade of CASP: progress, bottlenecks and prognosis in protein structure prediction.Curr Opin Struct Biol. 2005 Jun;15(3):285-9. doi: 10.1016/j.sbi.2005.05.011. Curr Opin Struct Biol. 2005. PMID: 15939584 Review.
-
Evolution of protein interactions: from interactomes to interfaces.Arch Biochem Biophys. 2014 Jul 15;554:65-75. doi: 10.1016/j.abb.2014.05.010. Epub 2014 May 20. Arch Biochem Biophys. 2014. PMID: 24853495 Review.
Cited by
-
Combining cysteine scanning with chemical labeling to map protein-protein interactions and infer bound structure in an intrinsically disordered region.Front Mol Biosci. 2022 Oct 7;9:997653. doi: 10.3389/fmolb.2022.997653. eCollection 2022. Front Mol Biosci. 2022. PMID: 36275627 Free PMC article.
-
Coevolutive, evolutive and stochastic information in protein-protein interactions.Comput Struct Biotechnol J. 2019 Nov 20;17:1429-1435. doi: 10.1016/j.csbj.2019.10.005. eCollection 2019. Comput Struct Biotechnol J. 2019. PMID: 31871588 Free PMC article.
-
An Ensemble Classifier to Predict Protein-Protein Interactions by Combining PSSM-based Evolutionary Information with Local Binary Pattern Model.Int J Mol Sci. 2019 Jul 17;20(14):3511. doi: 10.3390/ijms20143511. Int J Mol Sci. 2019. PMID: 31319578 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

