The role of myosin II in glioma invasion: A mathematical model
- PMID: 28166231
- PMCID: PMC5293275
- DOI: 10.1371/journal.pone.0171312
The role of myosin II in glioma invasion: A mathematical model
Abstract
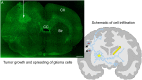
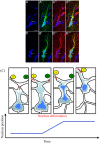
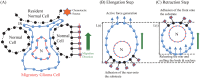
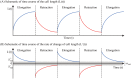
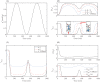
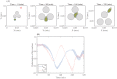
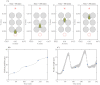
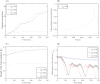
Gliomas are malignant tumors that are commonly observed in primary brain cancer. Glioma cells migrate through a dense network of normal cells in microenvironment and spread long distances within brain. In this paper we present a two-dimensional multiscale model in which a glioma cell is surrounded by normal cells and its migration is controlled by cell-mechanical components in the microenvironment via the regulation of myosin II in response to chemoattractants. Our simulation results show that the myosin II plays a key role in the deformation of the cell nucleus as the glioma cell passes through the narrow intercellular space smaller than its nuclear diameter. We also demonstrate that the coordination of biochemical and mechanical components within the cell enables a glioma cell to take the mode of amoeboid migration. This study sheds lights on the understanding of glioma infiltration through the narrow intercellular spaces and may provide a potential approach for the development of anti-invasion strategies via the injection of chemoattractants for localization.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures








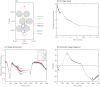



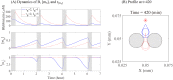



References
-
- Lund-Johansen M, Bjerkvig R, Humphrey PA, Bigner SH, Bigner DD, Laerum OD. Effect of epidermal growth factor on glioma cell growth, migration, and invasion in vitro. Cancer Res. 1990;50(18):6039–44. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

