Evidence for disulfide bonds in SR Protein Kinase 1 (SRPK1) that are required for activity and nuclear localization
- PMID: 28166275
- PMCID: PMC5293202
- DOI: 10.1371/journal.pone.0171328
Evidence for disulfide bonds in SR Protein Kinase 1 (SRPK1) that are required for activity and nuclear localization
Abstract
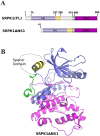
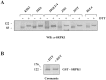
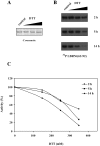
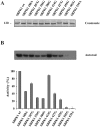
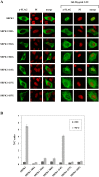
Serine/arginine protein kinases (SRPKs) phosphorylate Arg/Ser dipeptide-containing proteins that play crucial roles in a broad spectrum of basic cellular processes. The existence of a large internal spacer sequence that separates the bipartite kinase catalytic core is a unique structural feature of SRPKs. Previous structural studies on a catalytically active fragment of SRPK1, which lacks the main part of the spacer domain, revealed that SRPK1 remains in an active state without any post-translational modifications or specific intra-protein interactions, while the spacer domain is depicted as a loop structure, outside the kinase core. Using systematic mutagenesis we now provide evidence that replacement of any individual cysteine residue in the spacer, apart from Cys414, or in its proximal flaking ends of the two kinase catalytic domains has an impact on kinase activity. Furthermore, the cysteine residues are critical for nuclear translocation of SRPK1 in response to genotoxic stress and SRPK1-dependent splicing of a reporter gene. While replacement of Cys207, Cys502 and Cys539 of the catalytic domains is predicted to distort the kinase active structure, our findings suggest that Cys356, Cys386, Cys427 and Cys455 of the spacer domain and Cys188 of the first catalytic domain are engaged in disulfide bridging. We propose that such a network of intramolecular disulfide bonds mediates the bending of the spacer region thus allowing the proximal positioning of the two catalytic subunits which is a prerequisite for SRPK1 activity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Nuclear Translocation of SRPKs Is Associated with 5-FU and Cisplatin Sensitivity in HeLa and T24 Cells.Cells. 2021 Mar 30;10(4):759. doi: 10.3390/cells10040759. Cells. 2021. PMID: 33808326 Free PMC article.
-
Regulated cellular partitioning of SR protein-specific kinases in mammalian cells.Mol Biol Cell. 2006 Feb;17(2):876-85. doi: 10.1091/mbc.e05-10-0963. Epub 2005 Nov 30. Mol Biol Cell. 2006. PMID: 16319169 Free PMC article.
-
A conserved sequence motif bridges two protein kinases for enhanced phosphorylation and nuclear function of a splicing factor.FEBS J. 2021 Jan;288(2):566-581. doi: 10.1111/febs.15351. Epub 2020 May 23. FEBS J. 2021. PMID: 32359191
-
Phosphorylation mechanism and structure of serine-arginine protein kinases.FEBS J. 2011 Feb;278(4):587-97. doi: 10.1111/j.1742-4658.2010.07992.x. Epub 2011 Jan 12. FEBS J. 2011. PMID: 21205204 Free PMC article. Review.
-
Serine-arginine protein kinases: a small protein kinase family with a large cellular presence.FEBS J. 2011 Feb;278(4):570-86. doi: 10.1111/j.1742-4658.2010.07987.x. Epub 2011 Jan 12. FEBS J. 2011. PMID: 21205200 Review.
Cited by
-
Good Cop, Bad Cop: The Different Roles of SRPKs.Front Genet. 2022 Jun 2;13:902718. doi: 10.3389/fgene.2022.902718. eCollection 2022. Front Genet. 2022. PMID: 35719374 Free PMC article. Review.
-
Phosphorylation mediated regulation of RNA splicing in plants.Front Plant Sci. 2023 Sep 14;14:1249057. doi: 10.3389/fpls.2023.1249057. eCollection 2023. Front Plant Sci. 2023. PMID: 37780493 Free PMC article. Review.
-
Nuclear Translocation of SRPKs Is Associated with 5-FU and Cisplatin Sensitivity in HeLa and T24 Cells.Cells. 2021 Mar 30;10(4):759. doi: 10.3390/cells10040759. Cells. 2021. PMID: 33808326 Free PMC article.
-
Serine arginine protein kinase 1 (SRPK1): a moonlighting protein with theranostic ability in cancer prevention.Mol Biol Rep. 2019 Feb;46(1):1487-1497. doi: 10.1007/s11033-018-4545-5. Epub 2018 Dec 8. Mol Biol Rep. 2019. PMID: 30535769 Review.
-
SR Protein Kinase 1 Inhibition by TAF15.Cells. 2022 Dec 28;12(1):126. doi: 10.3390/cells12010126. Cells. 2022. PMID: 36611919 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

