Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer
- PMID: 28166537
- PMCID: PMC5448706
- DOI: 10.1038/nature21357
Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer
Abstract
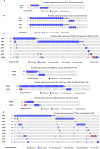
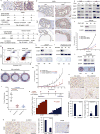
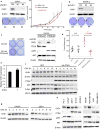
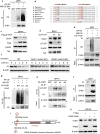
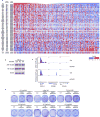
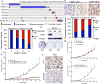
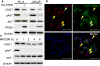
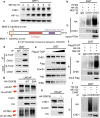
Synthetic lethality and collateral lethality are two well-validated conceptual strategies for identifying therapeutic targets in cancers with tumour-suppressor gene deletions. Here, we explore an approach to identify potential synthetic-lethal interactions by screening mutually exclusive deletion patterns in cancer genomes. We sought to identify 'synthetic-essential' genes: those that are occasionally deleted in some cancers but are almost always retained in the context of a specific tumour-suppressor deficiency. We also posited that such synthetic-essential genes would be therapeutic targets in cancers that harbour specific tumour-suppressor deficiencies. In addition to known synthetic-lethal interactions, this approach uncovered the chromatin helicase DNA-binding factor CHD1 as a putative synthetic-essential gene in PTEN-deficient cancers. In PTEN-deficient prostate and breast cancers, CHD1 depletion profoundly and specifically suppressed cell proliferation, cell survival and tumorigenic potential. Mechanistically, functional PTEN stimulates the GSK3β-mediated phosphorylation of CHD1 degron domains, which promotes CHD1 degradation via the β-TrCP-mediated ubiquitination-proteasome pathway. Conversely, PTEN deficiency results in stabilization of CHD1, which in turn engages the trimethyl lysine-4 histone H3 modification to activate transcription of the pro-tumorigenic TNF-NF-κB gene network. This study identifies a novel PTEN pathway in cancer and provides a framework for the discovery of 'trackable' targets in cancers that harbour specific tumour-suppressor deficiencies.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Comment in
-
Prostate cancer: Fatal interaction: a new target identified.Nat Rev Urol. 2017 May;14(5):258-259. doi: 10.1038/nrurol.2017.29. Epub 2017 Feb 21. Nat Rev Urol. 2017. PMID: 28248947 No abstract available.
-
Drug therapy: Exploiting synthetic lethality to improve cancer therapy.Nat Rev Clin Oncol. 2017 Jun;14(6):331-332. doi: 10.1038/nrclinonc.2017.46. Epub 2017 Mar 29. Nat Rev Clin Oncol. 2017. PMID: 28352131 No abstract available.
References
-
- Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278:1064–1068. - PubMed
-
- Cairns P, et al. Frequent inactivation of PTEN/MMAC1 in primary prostate cancer. Cancer Res. 1997;57:4997–5000. - PubMed
-
- Wang S, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer cell. 2003;4:209–221. doi:S1535610803002150 [pii] - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

