Identification and expression profile analysis of odorant binding protein and chemosensory protein genes in Bemisia tabaci MED by head transcriptome
- PMID: 28166541
- PMCID: PMC5293548
- DOI: 10.1371/journal.pone.0171739
Identification and expression profile analysis of odorant binding protein and chemosensory protein genes in Bemisia tabaci MED by head transcriptome
Abstract
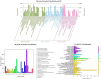
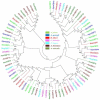
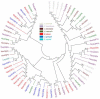
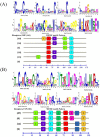
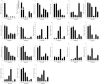
Odorant binding proteins (OBPs) and chemosensory proteins (CSPs) of arthropods are thought to be involved in chemical recognition which regulates pivotal behaviors including host choice, copulation and reproduction. In insects, OBPs and CSPs located mainly in the antenna but they have not been systematically characterized yet in Bemisia tabaci which is a cryptic species complex and could damage more than 600 plant species. In this study, among the 106,893 transcripts in the head assembly, 8 OBPs and 13 CSPs were identified in B. tabaci MED based on head transcriptomes of adults. Phylogenetic analyses were conducted to investigate the relationships of B. tabaci OBPs and CSPs with those from several other important Hemipteran species, and the motif-patterns between Hemiptera OBPs and CSPs were also compared by MEME. The expression profiles of the OBP and CSP genes in different tissues of B. tabaci MED adults were analyzed by real-time qPCR. Seven out of the 8 OBPs found in B. tabaci MED were highly expressed in the head. Conversely, only 4 CSPs were enriched in the head, while the other nine CSPs were specifically expressed in other tissues. Our findings pave the way for future research on chemical recognition of B. tabaci at the molecular level.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

