Proceedings of the 15th Annual UT-KBRIN Bioinformatics Summit 2016 : Cadiz, KY, USA. 8-10 April 2016
- PMID: 28166713
- PMCID: PMC5001208
- DOI: 10.1186/s12859-016-1154-y
Proceedings of the 15th Annual UT-KBRIN Bioinformatics Summit 2016 : Cadiz, KY, USA. 8-10 April 2016
Abstract
I1 Proceedings of the Fifteenth Annual UT- KBRIN Bioinformatics Summit 2016
Eric C. Rouchka, Julia H. Chariker, Benjamin J. Harrison, Juw Won Park
P1 CC-PROMISE: Projection onto the Most Interesting Statistical Evidence (PROMISE) with Canonical Correlation to integrate gene expression and methylation data with multiple pharmacologic and clinical endpoints
Xueyuan Cao, Stanley Pounds, Susana Raimondi, James Downing, Raul Ribeiro, Jeffery Rubnitz, Jatinder Lamba
P2 Integration of microRNA-mRNA interaction networks with gene expression data to increase experimental power
Bernie J Daigle, Jr.
P3 Designing and writing software for in silico subtractive hybridization of large eukaryotic genomes
Deborah Burgess, Stephanie Gehrlich, John C Carmen
P4 Tracking the molecular evolution of Pax gene
Nicholas Johnson; Chandrakanth Emani
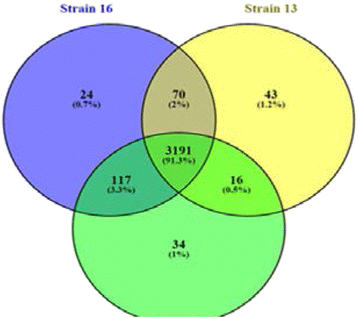
P5 Identifying genetic differences in thermally dimorphic and state specific fungi using in silico genomic comparison
Stephanie Gehrlich, Deborah Burgess, John C Carmen
P6 Identification of conserved genomic regions and variation therein amongst Cetartiodactyla species using next generation sequencing
Kalpani De Silva, Michael P Heaton, Theodore S Kalbfleisch
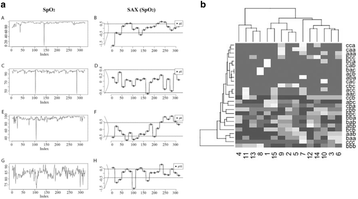
P7 Mining physiological data to identify patients with similar medical events and phenotypes
Teeradache Viangteeravat, Rahul Mudunuri, Oluwaseun Ajayi, Fatih Şen, Eunice Y Huang
P8 Smart brief for home health monitoring
Mohammad Mohebbi, Luaire Florian, Douglas J Jackson, John F Naber
P9 Side-effect term matching for computational adverse drug reaction predictions
AKM Sabbir, Sally R Ellingson
P10 Enrichment vs robustness: A comparison of transcriptomic data clustering metrics
Yuping Lu, Charles A Phillips, Michael A Langston
P11 Deep neural networks for transcriptome-based cancer classification
Rahul K Sevakula, Raghuveer Thirukovalluru, Nishchal K. Verma, Yan Cui
P12 Motif discovery using K-means clustering
Mohammed Sayed, Juw Won Park
P13 Large scale discovery of active enhancers from nascent RNA sequencing
Jing Wang, Qi Liu, Yu Shyr
P14 Computationally characterizing genomic pipelines and benchmarking results using GATK best practices on the high performance computing cluster at the University of Kentucky
Xiaofei Zhang, Sally R Ellingson
P15 Development of approaches enabling the identification of abnormal gene expression from RNA-Seq in personalized oncology
Naresh Prodduturi, Gavin R Oliver, Diane Grill, Jie Na, Jeanette Eckel-Passow, Eric W Klee
P16 Processing RNA-Seq data of plants infected with coffee ringspot virus
Michael M Goodin, Mark Farman, Harrison Inocencio, Chanyong Jang, Jerzy W Jaromczyk, Neil Moore, Kelly Sovacool
P17 Comparative transcriptomics of three Acinetobacter baumanii clinical isolates with different antibiotic resistance patterns
Leon Dent, Mike Izban, Sammed Mandape, Shruti Sakhare, Siddharth Pratap, Dana Marshall
P18 Metagenomic assessment of possible microbial contamination in the equine reference genome assembly
M Scotty DePriest, James N MacLeod, Theodore S Kalbfleisch
P19 Molecular evolution of cancer driver genes
Chandrakanth Emani, Hanady Adam, Ethan Blandford, Joel Campbell, Joshua Castlen, Brittany Dixon, Ginger Gilbert, Aaron Hall, Philip Kreisle, Jessica Lasher, Bethany Oakes, Allison Speer, Maximilian Valentine
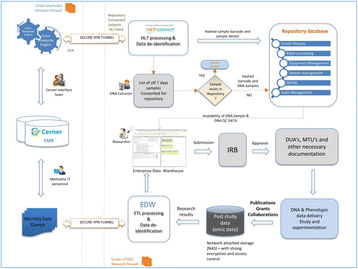
P20 Biorepository Laboratory Information Management System
Naga Satya V Rao Nagisetty, Rony Jose, Teeradache Viangteeravat, Robert Rooney, David Hains
Figures
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

