bModelTest: Bayesian phylogenetic site model averaging and model comparison
- PMID: 28166715
- PMCID: PMC5294809
- DOI: 10.1186/s12862-017-0890-6
bModelTest: Bayesian phylogenetic site model averaging and model comparison
Abstract
Background: Reconstructing phylogenies through Bayesian methods has many benefits, which include providing a mathematically sound framework, providing realistic estimates of uncertainty and being able to incorporate different sources of information based on formal principles. Bayesian phylogenetic analyses are popular for interpreting nucleotide sequence data, however for such studies one needs to specify a site model and associated substitution model. Often, the parameters of the site model is of no interest and an ad-hoc or additional likelihood based analysis is used to select a single site model.
Results: bModelTest allows for a Bayesian approach to inferring and marginalizing site models in a phylogenetic analysis. It is based on trans-dimensional Markov chain Monte Carlo (MCMC) proposals that allow switching between substitution models as well as estimating the posterior probability for gamma-distributed rate heterogeneity, a proportion of invariable sites and unequal base frequencies. The model can be used with the full set of time-reversible models on nucleotides, but we also introduce and demonstrate the use of two subsets of time-reversible substitution models.
Conclusion: With the new method the site model can be inferred (and marginalized) during the MCMC analysis and does not need to be pre-determined, as is now often the case in practice, by likelihood-based methods. The method is implemented in the bModelTest package of the popular BEAST 2 software, which is open source, licensed under the GNU Lesser General Public License and allows joint site model and tree inference under a wide range of models.
Keywords: Model averaging; Model comparison; Model selection; ModelTest; Phylogenetic model averaging; Phylogenetic model comparison; Site model; Statistical phylogenetics; Substitution model.
Figures
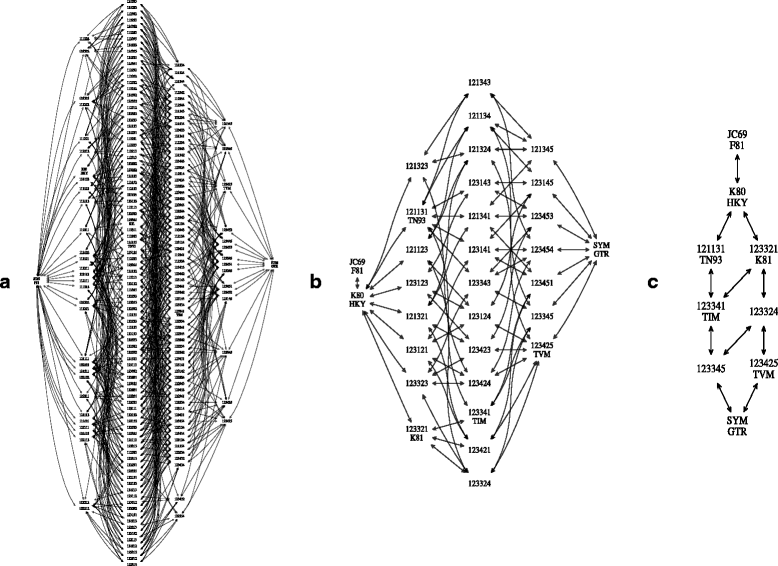
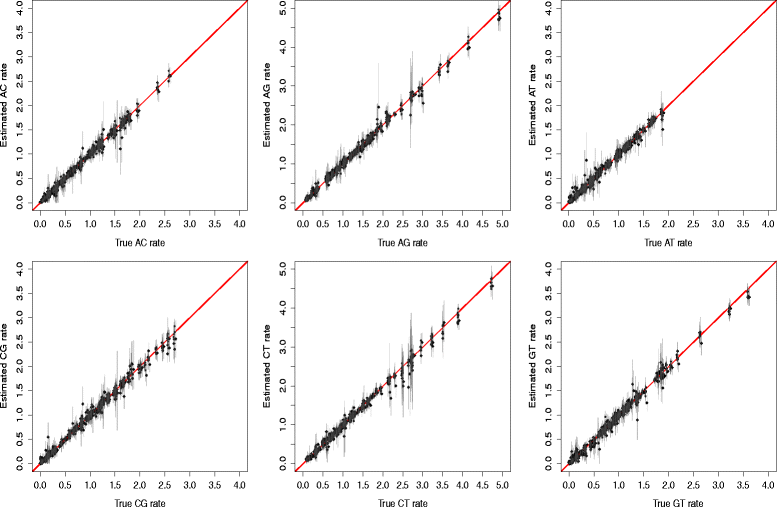
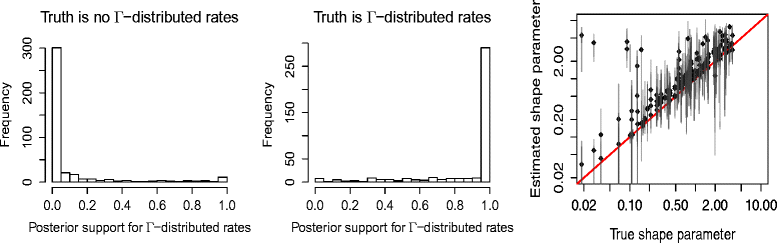

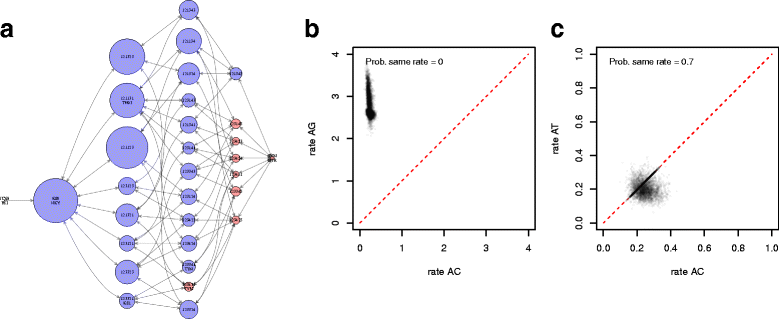
References
-
- Gu X, Fu YX, Li WH. Maximum likelihood estimation of the heterogeneity of substitution rate among nucleotide sites. Mol Biol Evol. 1995;12(4):546–7. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

