Toll-like receptor 3 activation selectively reverses HIV latency in microglial cells
- PMID: 28166799
- PMCID: PMC5294768
- DOI: 10.1186/s12977-017-0335-8
Toll-like receptor 3 activation selectively reverses HIV latency in microglial cells
Abstract
Background: Multiple toll-like receptors (TLRs) are expressed in cells of the monocytic lineage, including microglia, which constitute the major reservoir for human immunodeficiency virus (HIV) infection in the brain. We hypothesized that TLR receptor mediated responses to inflammatory conditions by microglial cells in the central nervous system (CNS) are able to induce latent HIV proviruses, and contribute to the etiology of HIV-associated neurocognitive disorders.
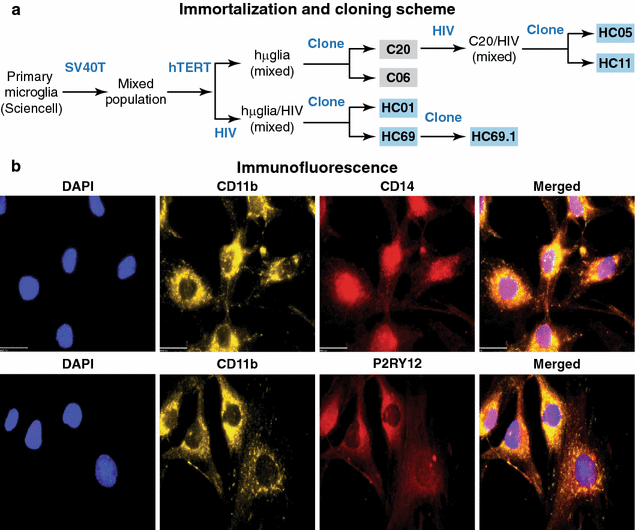
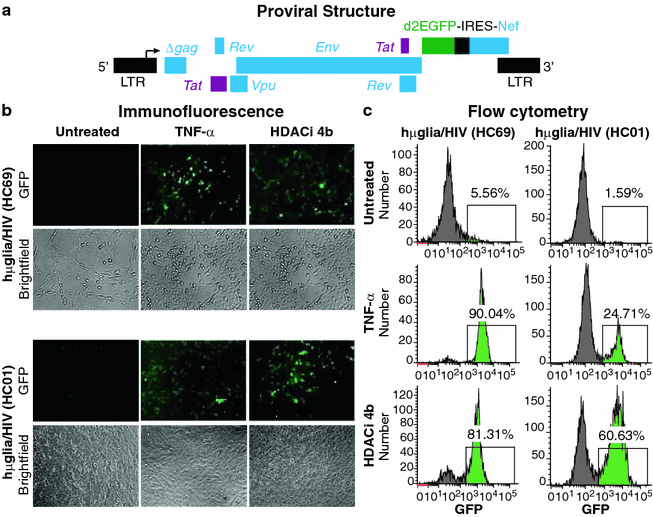
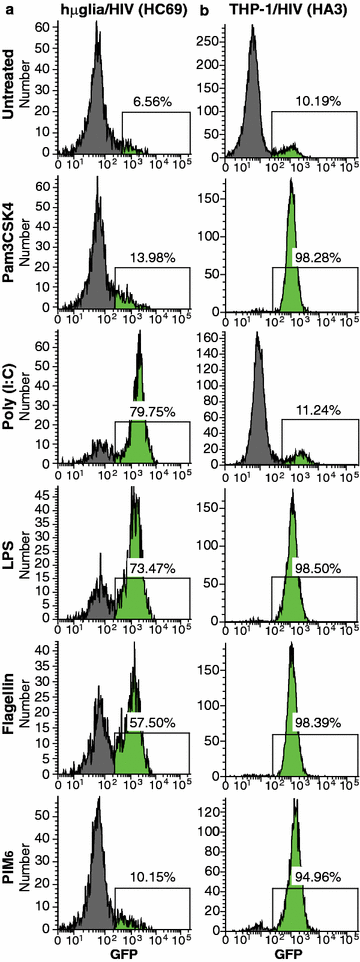
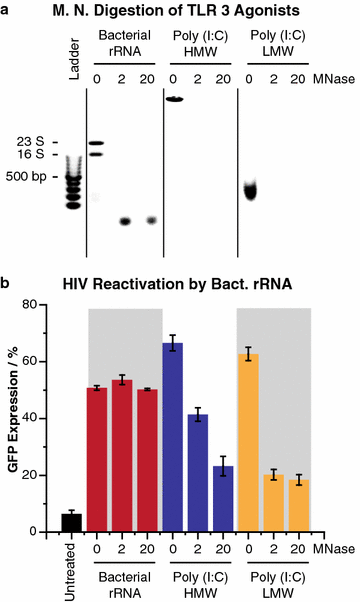
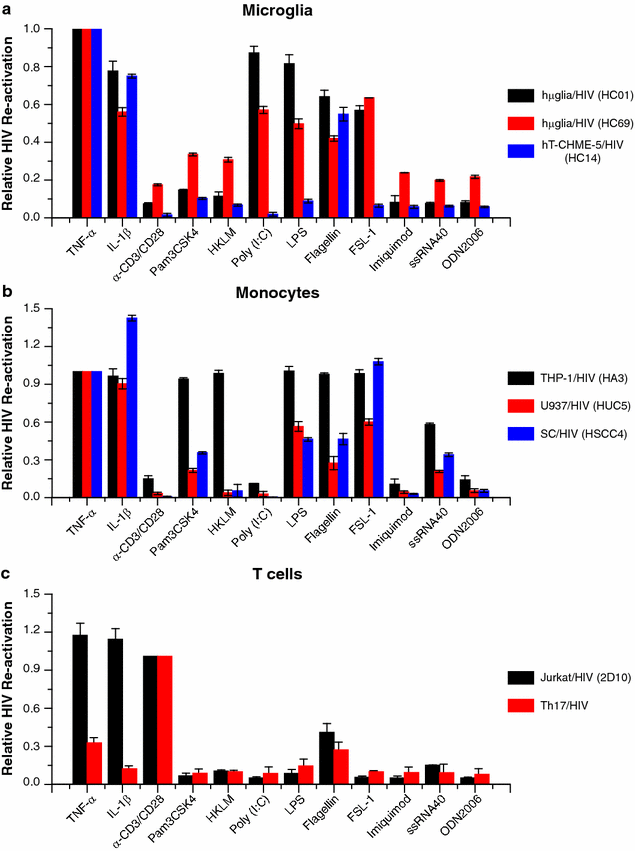
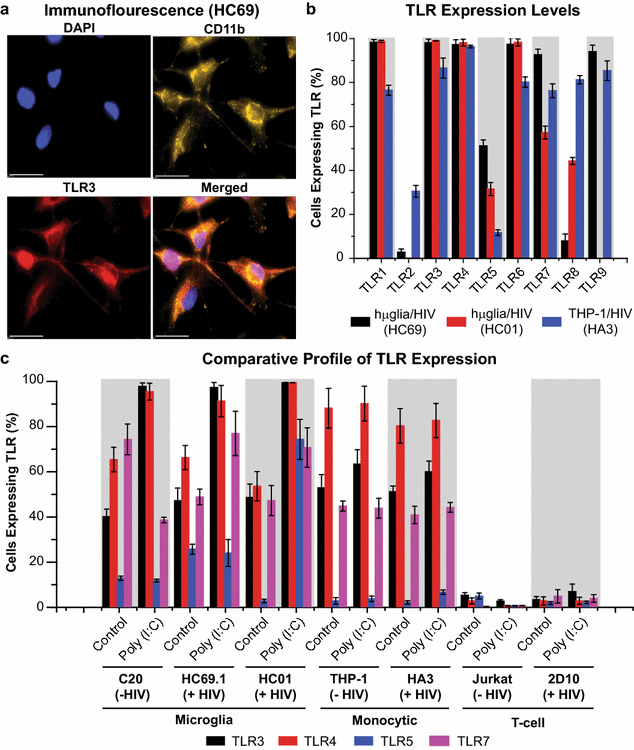
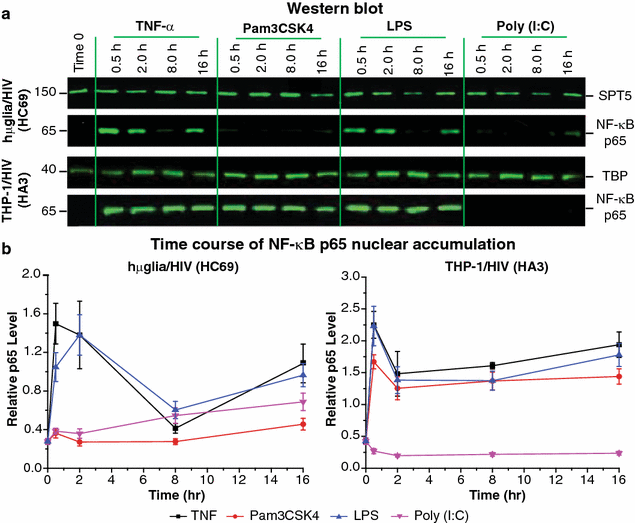
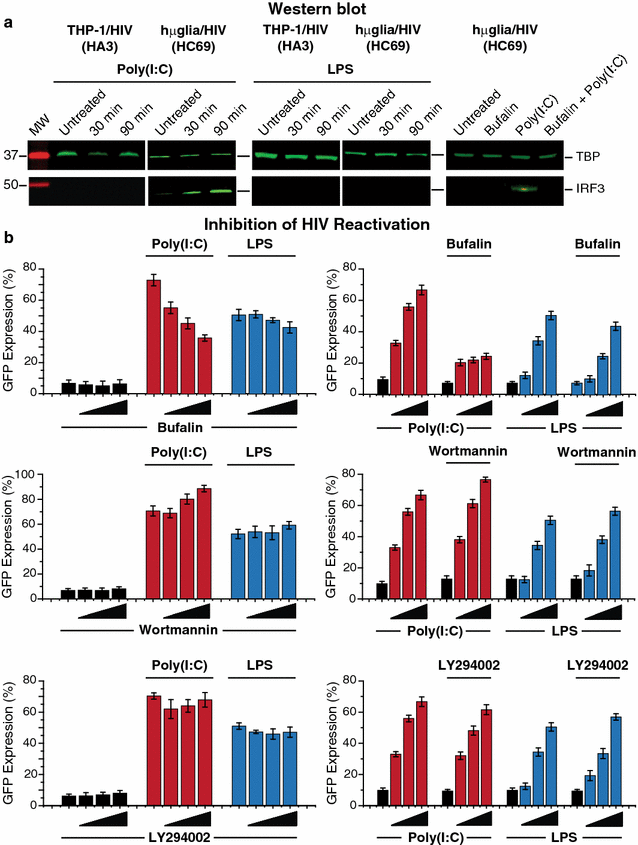
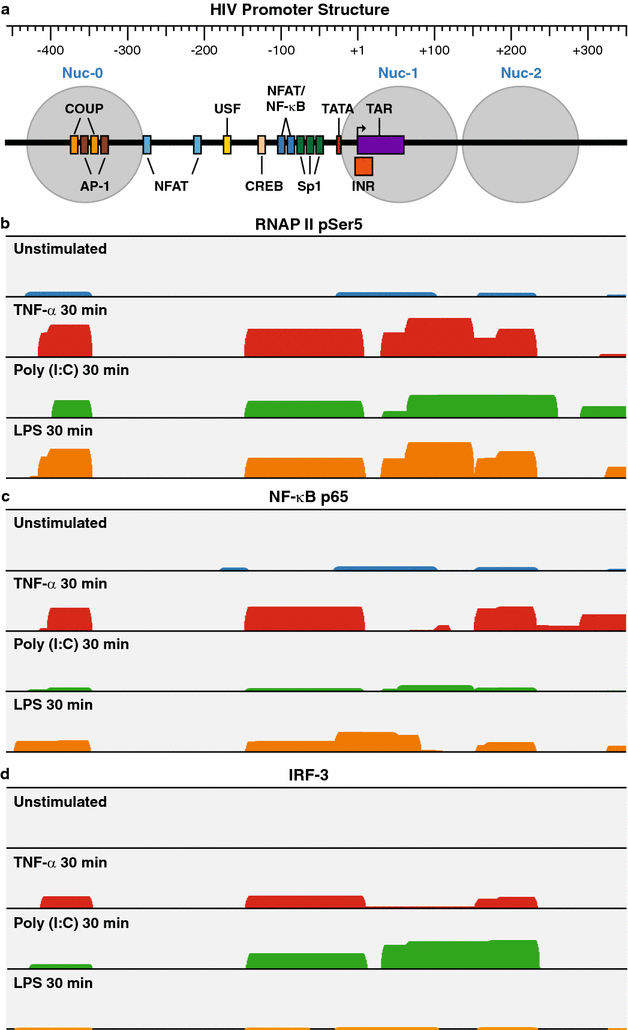
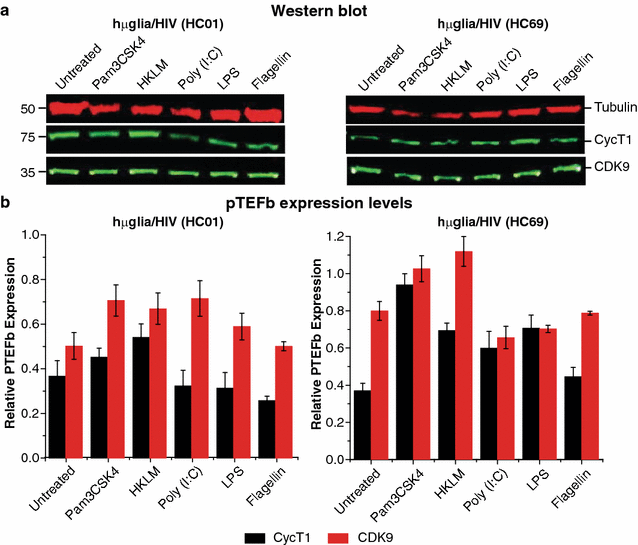
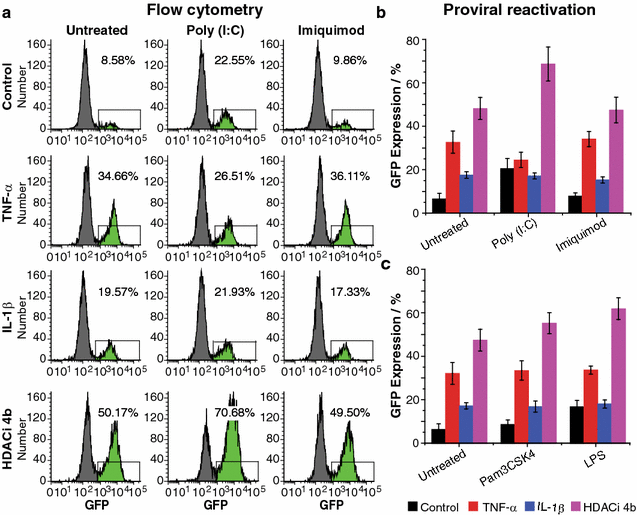
Results: Newly developed human microglial cell lines (hµglia), obtained by immortalizing human primary microglia with simian virus-40 (SV40) large T antigen and the human telomerase reverse transcriptase, were used to generate latently infected cells using a single-round HIV virus carrying a green fluorescence protein reporter (hµglia/HIV, clones HC01 and HC69). Treatment of these cells with a panel of TLR ligands showed surprisingly that two potent TLR3 agonists, poly (I:C) and bacterial ribosomal RNA potently reactivated HIV in hμglia/HIV cells. LPS (TLR4 agonist), flagellin (TLR5 agonist), and FSL-1 (TLR6 agonist) reactivated HIV to a lesser extent, while Pam3CSK4 (TLR2/1 agonist) and HKLM (TLR2 agonist) only weakly reversed HIV latency in these cells. While agonists for TLR2/1, 4, 5 and 6 reactivated HIV through transient NF-κB induction, poly (I:C), the TLR3 agonist, did not activate NF-κB, and instead induced the virus by a previously unreported mechanism mediated by IRF3. The selective induction of IRF3 by poly (I:C) was confirmed by chromatin immunoprecipitation (ChIP) analysis. In comparison, in latently infected rat-derived microglial cells (hT-CHME-5/HIV, clone HC14), poly (I:C), LPS and flagellin were only partially active. The TLR response profile in human microglial cells is also distinct from that shown by latently infected monocyte cell lines (THP-1/HIV, clone HA3, U937/HIV, clone HUC5, and SC/HIV, clone HSCC4), where TLR2/1, 4, 5, 6 or 8, but not for TLR3, 7 or 9, reactivated HIV.
Conclusions: TLR signaling, in particular TLR3 activation, can efficiently reactivate HIV transcription in infected microglia, but not in monocytes or T cells. The unique response profile of microglial cells to TLR3 is fundamental to understanding how the virus responds to continuous microbial exposure, especially during inflammatory episodes, that characterizes HIV infection in the CNS.
Keywords: HIV latency; HIV-associated neurocognitive disorders; Microglial cells; TLR3; Toll-like receptors.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

