Regulation of Polyhydroxybutyrate Accumulation in Sinorhizobium meliloti by the Trans-Encoded Small RNA MmgR
- PMID: 28167519
- PMCID: PMC5370414
- DOI: 10.1128/JB.00776-16
Regulation of Polyhydroxybutyrate Accumulation in Sinorhizobium meliloti by the Trans-Encoded Small RNA MmgR
Abstract
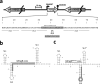
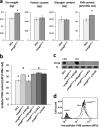
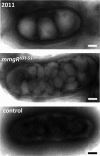
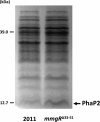
Riboregulation has a major role in the fine-tuning of multiple bacterial processes. Among the RNA players, trans-encoded untranslated small RNAs (sRNAs) regulate complex metabolic networks by tuning expression from multiple target genes in response to numerous signals. In Sinorhizobium meliloti, over 400 sRNAs are expressed under different stimuli. The sRNA MmgR (standing for
Keywords: MmgR; PHB; Sinorhizobium meliloti; riboregulation; small RNA.
Copyright © 2017 American Society for Microbiology.
Figures



Similar articles
-
Expression of the small regulatory RNA gene mmgR is regulated negatively by AniA and positively by NtrC in Sinorhizobium meliloti 2011.Microbiology (Reading). 2018 Jan;164(1):88-98. doi: 10.1099/mic.0.000586. Epub 2017 Dec 7. Microbiology (Reading). 2018. PMID: 29214973
-
Phylogenetic distribution and evolutionary pattern of an α-proteobacterial small RNA gene that controls polyhydroxybutyrate accumulation in Sinorhizobium meliloti.Mol Phylogenet Evol. 2016 Jun;99:182-193. doi: 10.1016/j.ympev.2016.03.026. Epub 2016 Mar 24. Mol Phylogenet Evol. 2016. PMID: 27033949
-
Expression of the Sinorhizobium meliloti small RNA gene mmgR is controlled by the nitrogen source.FEMS Microbiol Lett. 2016 May;363(9):fnw069. doi: 10.1093/femsle/fnw069. Epub 2016 Mar 23. FEMS Microbiol Lett. 2016. PMID: 27010014
-
Insights into the noncoding RNome of nitrogen-fixing endosymbiotic α-proteobacteria.Mol Plant Microbe Interact. 2013 Feb;26(2):160-7. doi: 10.1094/MPMI-07-12-0186-CR. Mol Plant Microbe Interact. 2013. PMID: 22991999 Review.
-
Riboregulation in bacteria: From general principles to novel mechanisms of the trp attenuator and its sRNA and peptide products.Wiley Interdiscip Rev RNA. 2022 May;13(3):e1696. doi: 10.1002/wrna.1696. Epub 2021 Oct 14. Wiley Interdiscip Rev RNA. 2022. PMID: 34651439 Review.
Cited by
-
Biotechnological Approaches for Enhancing Polyhydroxyalkanoates (PHAs) Production: Current and Future Perspectives.Curr Microbiol. 2023 Sep 20;80(11):345. doi: 10.1007/s00284-023-03452-4. Curr Microbiol. 2023. PMID: 37731015 Review.
-
A Genome-Wide Prediction and Identification of Intergenic Small RNAs by Comparative Analysis in Mesorhizobium huakuii 7653R.Front Microbiol. 2017 Sep 8;8:1730. doi: 10.3389/fmicb.2017.01730. eCollection 2017. Front Microbiol. 2017. PMID: 28943874 Free PMC article.
-
Post-Transcriptional Control in the Regulation of Polyhydroxyalkanoates Synthesis.Life (Basel). 2021 Aug 20;11(8):853. doi: 10.3390/life11080853. Life (Basel). 2021. PMID: 34440597 Free PMC article. Review.
-
Synthetase of the methyl donor S-adenosylmethionine from nitrogen-fixing α-rhizobia can bind functionally diverse RNA species.RNA Biol. 2021 Aug;18(8):1111-1123. doi: 10.1080/15476286.2020.1829365. Epub 2020 Oct 10. RNA Biol. 2021. PMID: 33043803 Free PMC article.
-
Quantification of Bacterial Polyhydroxybutyrate Content by Flow Cytometry.Bio Protoc. 2017 Dec 5;7(23):e2638. doi: 10.21769/BioProtoc.2638. eCollection 2017 Dec 5. Bio Protoc. 2017. PMID: 34595305 Free PMC article.
References
-
- Horvath B, Kondorosi E, John M, Schmidt J, Torok I, Gyorgypal Z, Barabas I, Wieneke U, Schell J, Kondorosi A. 1986. Organization, structure and symbiotic function of Rhizobium meliloti nodulation genes determining host specificity for alfalfa. Cell 46:335–343. doi:10.1016/0092-8674(86)90654-9. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

