A New Essential Cell Division Protein in Caulobacter crescentus
- PMID: 28167520
- PMCID: PMC5370419
- DOI: 10.1128/JB.00811-16
A New Essential Cell Division Protein in Caulobacter crescentus
Abstract
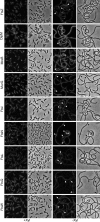
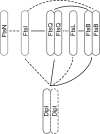
Bacterial cell division is a complex process that relies on a multiprotein complex composed of a core of widely conserved and generally essential proteins and on accessory proteins that vary in number and identity in different bacteria. The assembly of this complex and, particularly, the initiation of constriction are regulated processes that have come under intensive study. In this work, we characterize the function of DipI, a protein conserved in Alphaproteobacteria and Betaproteobacteria that is essential in Caulobacter crescentus Our results show that DipI is a periplasmic protein that is recruited late to the division site and that it is required for the initiation of constriction. The recruitment of the conserved cell division proteins is not affected by the absence of DipI, but localization of DipI to the division site occurs only after a mature divisome has formed. Yeast two-hybrid analysis showed that DipI strongly interacts with the FtsQLB complex, which has been recently implicated in regulating constriction initiation. A possible role of DipI in this process is discussed.IMPORTANCE Bacterial cell division is a complex process for which most bacterial cells assemble a multiprotein complex that consists of conserved proteins and of accessory proteins that differ among bacterial groups. In this work, we describe a new cell division protein (DipI) present only in a group of bacteria but essential in Caulobacter crescentus Cells devoid of DipI cannot constrict. Although a mature divisome is required for DipI recruitment, DipI is not needed for recruiting other division proteins. These results, together with the interaction of DipI with a protein complex that has been suggested to regulate cell wall synthesis during division, suggest that DipI may be part of the regulatory mechanism that controls constriction initiation.
Keywords: Caulobacter crescentus; SH3 domain; bacterial cell division; constriction initiation; divisome.
Copyright © 2017 American Society for Microbiology.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

