The Crystal Structure of Cytochrome P450 4B1 (CYP4B1) Monooxygenase Complexed with Octane Discloses Several Structural Adaptations for ω-Hydroxylation
- PMID: 28167536
- PMCID: PMC5392703
- DOI: 10.1074/jbc.M117.775494
The Crystal Structure of Cytochrome P450 4B1 (CYP4B1) Monooxygenase Complexed with Octane Discloses Several Structural Adaptations for ω-Hydroxylation
Abstract
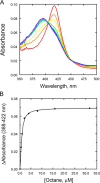
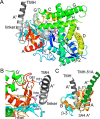
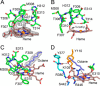
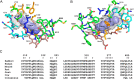
P450 family 4 fatty acid ω-hydroxylases preferentially oxygenate primary C-H bonds over adjacent, energetically favored secondary C-H bonds, but the mechanism explaining this intriguing preference is unclear. To this end, the structure of rabbit P450 4B1 complexed with its substrate octane was determined by X-ray crystallography to define features of the active site that contribute to a preference for ω-hydroxylation. The structure indicated that octane is bound in a narrow active-site cavity that limits access of the secondary C-H bond to the reactive intermediate. A highly conserved sequence motif on helix I contributes to positioning the terminal carbon of octane for ω-hydroxylation. Glu-310 of this motif auto-catalytically forms an ester bond with the heme 5-methyl, and the immobilized Glu-310 contributes to substrate positioning. The preference for ω-hydroxylation was decreased in an E310A mutant having a shorter side chain, but the overall rates of metabolism were retained. E310D and E310Q substitutions having longer side chains exhibit lower overall rates, likely due to higher conformational entropy for these residues, but they retained high preferences for octane ω-hydroxylation. Sequence comparisons indicated that active-site residues constraining octane for ω-hydroxylation are conserved in family 4 P450s. Moreover, the heme 7-propionate is positioned in the active site and provides additional restraints on substrate binding. In conclusion, P450 4B1 exhibits structural adaptations for ω-hydroxylation that include changes in the conformation of the heme and changes in a highly conserved helix I motif that is associated with selective oxygenation of unactivated primary C-H bonds.
Keywords: X-ray crystallography; cytochrome P450; enzyme structure; fatty acid metabolism; heme; membrane protein; xenobiotic.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

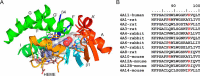
Similar articles
-
Noncovalent interactions dominate dynamic heme distortion in cytochrome P450 4B1.J Biol Chem. 2018 Jul 20;293(29):11433-11446. doi: 10.1074/jbc.RA118.004044. Epub 2018 Jun 1. J Biol Chem. 2018. PMID: 29858244 Free PMC article.
-
ω- versus (ω-1)-hydroxylation: Cytochrome P450 4B1 sterics make the call.J Biol Chem. 2017 Mar 31;292(13):5622-5623. doi: 10.1074/jbc.H117.775494. J Biol Chem. 2017. PMID: 28363936 Free PMC article.
-
Mechanism and role of covalent heme binding in the CYP4 family of P450 enzymes and the mammalian peroxidases.Drug Metab Rev. 2008;40(3):405-26. doi: 10.1080/03602530802186439. Drug Metab Rev. 2008. PMID: 18642140 Review.
-
Identification of a meander region proline residue critical for heme binding to cytochrome P450: implications for the catalytic function of human CYP4B1.Biochemistry. 1998 Sep 15;37(37):12847-51. doi: 10.1021/bi981280m. Biochemistry. 1998. PMID: 9737862
-
Structural control of cytochrome P450-catalyzed ω-hydroxylation.Arch Biochem Biophys. 2011 Mar 1;507(1):86-94. doi: 10.1016/j.abb.2010.08.011. Epub 2010 Aug 19. Arch Biochem Biophys. 2011. PMID: 20727847 Free PMC article. Review.
Cited by
-
Applications of protein engineering in the microbial synthesis of plant triterpenoids.Synth Syst Biotechnol. 2022 Oct 25;8(1):20-32. doi: 10.1016/j.synbio.2022.10.001. eCollection 2023 Mar. Synth Syst Biotechnol. 2022. PMID: 36381964 Free PMC article. Review.
-
Cytochrome P450: Implications for human breast cancer.Oncol Lett. 2021 Jul;22(1):548. doi: 10.3892/ol.2021.12809. Epub 2021 May 24. Oncol Lett. 2021. PMID: 34093769 Free PMC article. Review.
-
Noncovalent interactions dominate dynamic heme distortion in cytochrome P450 4B1.J Biol Chem. 2018 Jul 20;293(29):11433-11446. doi: 10.1074/jbc.RA118.004044. Epub 2018 Jun 1. J Biol Chem. 2018. PMID: 29858244 Free PMC article.
-
The Biosynthesis of Enzymatically Oxidized Lipids.Front Endocrinol (Lausanne). 2020 Nov 19;11:591819. doi: 10.3389/fendo.2020.591819. eCollection 2020. Front Endocrinol (Lausanne). 2020. PMID: 33329396 Free PMC article. Review.
-
A new naphthalene-based fluorogenic substrate for cytochrome P450 4A11.Biochem J. 2025 Jun 17;482(12):839-57. doi: 10.1042/BCJ20253130. Biochem J. 2025. PMID: 40454940 Free PMC article.
References
-
- Hsu M. H., Savas U., Griffin K. J., and Johnson E. F. (2007) Human cytochrome p450 family 4 enzymes: function, genetic variation and regulation. Drug. Metab. Rev. 39, 515–538 - PubMed
-
- Hardwick J. P. (2008) Cytochrome P450 ω-hydroxylase (CYP4) function in fatty acid metabolism and metabolic diseases. Biochem. Pharmacol. 75, 2263–2275 - PubMed
-
- Mortensen P. B., and Gregersen N. (1981) The biological origin of ketotic dicarboxylic aciduria: in vivo and in vitro investigations of the ω-oxidation of C6–C16-monocarboxylic acids in unstarved, starved and diabetic rats. Biochim. Biophys. Acta 666, 394–404 - PubMed
-
- Kam W., Kumaran K., and Landau B. R. (1978) Contribution of ω-oxidation to fatty acid oxidation by liver of rat and monkey. J. Lipid Res. 19, 591–600 - PubMed
-
- Björkhem I. (1978) On the quantitative importance of ω-oxidation of fatty acids. J. Lipid Res. 19, 585–590 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

