Molecular basis of fatty acid selectivity in the zDHHC family of S-acyltransferases revealed by click chemistry
- PMID: 28167757
- PMCID: PMC5338407
- DOI: 10.1073/pnas.1612254114
Molecular basis of fatty acid selectivity in the zDHHC family of S-acyltransferases revealed by click chemistry
Abstract
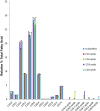
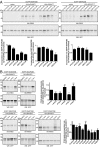
S-acylation is a major posttranslational modification, catalyzed by the zinc finger DHHC domain containing (zDHHC) enzyme family. S-acylated proteins can be modified by different fatty acids; however, very little is known about how zDHHC enzymes contribute to acyl chain heterogeneity. Here, we used fatty acid-azide/alkyne labeling of mammalian cells, showing their transformation into acyl-CoAs and subsequent click chemistry-based detection, to demonstrate that zDHHC enzymes have marked differences in their fatty acid selectivity. This difference in selectivity was apparent even for highly related enzymes, such as zDHHC3 and zDHHC7, which displayed a marked difference in their ability to use C18:0 acyl-CoA as a substrate. Furthermore, we identified isoleucine-182 in transmembrane domain 3 of zDHHC3 as a key determinant in limiting the use of longer chain acyl-CoAs by this enzyme. This study uncovered differences in the fatty acid selectivity profiles of cellular zDHHC enzymes and mapped molecular determinants governing this selectivity.
Keywords: S-acylation; acyl CoA; click chemistry; palmitoylation; zDHHC enzyme.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Smotrys JE, Linder ME. Palmitoylation of intracellular signaling proteins: Regulation and function. Annu Rev Biochem. 2004;73(1):559–587. - PubMed
-
- Linder ME, Deschenes RJ. Palmitoylation: Policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8(1):74–84. - PubMed
-
- Greaves J, Chamberlain LH. DHHC palmitoyl transferases: Substrate interactions and (patho)physiology. Trends Biochem Sci. 2011;36(5):245–253. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

