Dnmt3a restrains mast cell inflammatory responses
- PMID: 28167789
- PMCID: PMC5338402
- DOI: 10.1073/pnas.1616420114
Dnmt3a restrains mast cell inflammatory responses
Abstract
DNA methylation and specifically the DNA methyltransferase enzyme DNMT3A are involved in the pathogenesis of a variety of hematological diseases and in regulating the function of immune cells. Although altered DNA methylation patterns and mutations in DNMT3A correlate with mast cell proliferative disorders in humans, the role of DNA methylation in mast cell biology is not understood. By using mast cells lacking Dnmt3a, we found that this enzyme is involved in restraining mast cell responses to acute and chronic stimuli, both in vitro and in vivo. The exacerbated mast cell responses observed in the absence of Dnmt3a were recapitulated or enhanced by treatment with the demethylating agent 5-aza-2'-deoxycytidine as well as by down-modulation of Dnmt1 expression, further supporting the role of DNA methylation in regulating mast cell activation. Mechanistically, these effects were in part mediated by the dysregulated expression of the scaffold protein IQGAP2, which is characterized by the ability to regulate a wide variety of biological processes. Altogether, our data demonstrate that DNMT3A and DNA methylation are key modulators of mast cell responsiveness to acute and chronic stimulation.
Keywords: DNA methylation; epigenetics; inflammation; mast cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

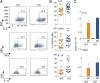
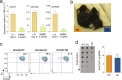

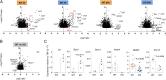
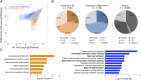



References
-
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. - PubMed
-
- Schübeler D. Function and information content of DNA methylation. Nature. 2015;517(7534):321–326. - PubMed
-
- Leoni C, Vincenzetti L, Emming S, Monticelli S. Epigenetics of T lymphocytes in health and disease. Swiss Med Wkly. 2015;145:w14191. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

