Estrogen receptor α wields treatment-specific enhancers between morphologically similar endometrial tumors
- PMID: 28167798
- PMCID: PMC5338442
- DOI: 10.1073/pnas.1615233114
Estrogen receptor α wields treatment-specific enhancers between morphologically similar endometrial tumors
Erratum in
-
Correction for Droog et al., Estrogen receptor α wields treatment-specific enhancers between morphologically similar endometrial tumors.Proc Natl Acad Sci U S A. 2018 Jan 30;115(5):E1075. doi: 10.1073/pnas.1800091115. Epub 2018 Jan 22. Proc Natl Acad Sci U S A. 2018. PMID: 29358402 Free PMC article. No abstract available.
Abstract
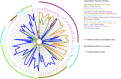
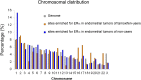
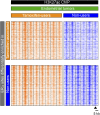
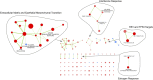
The DNA-binding sites of estrogen receptor α (ERα) show great plasticity under the control of hormones and endocrine therapy. Tamoxifen is a widely applied therapy in breast cancer that affects ERα interactions with coregulators and shifts the DNA-binding signature of ERα upon prolonged exposure in breast cancer. Although tamoxifen inhibits the progression of breast cancer, it increases the risk of endometrial cancer in postmenopausal women. We therefore asked whether the DNA-binding signature of ERα differs between endometrial tumors that arise in the presence or absence of tamoxifen, indicating divergent enhancer activity for tumors that develop in different endocrine milieus. Using ChIP sequencing (ChIP-seq), we compared the ERα profiles of 10 endometrial tumors from tamoxifen users with those of six endometrial tumors from nonusers and integrated these results with the transcriptomic data of 47 endometrial tumors from tamoxifen users and 64 endometrial tumors from nonusers. The ERα-binding sites in tamoxifen-associated endometrial tumors differed from those in the tumors from nonusers and had distinct underlying DNA sequences and divergent enhancer activity as marked by histone 3 containing the acetylated lysine 27 (H3K27ac). Because tamoxifen acts as an agonist in the postmenopausal endometrium, similar to estrogen in the breast, we compared ERα sites in tamoxifen-associated endometrial cancers with publicly available ERα ChIP-seq data in breast tumors and found a striking resemblance in the ERα patterns of the two tissue types. Our study highlights the divergence between endometrial tumors that arise in different hormonal conditions and shows that ERα enhancer use in human cancer differs in the presence of nonphysiological endocrine stimuli.
Keywords: ChIP-seq; breast cancer; endometrial cancer; estrogen receptor; tamoxifen.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Shiau AK, et al. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95(7):927–937. - PubMed
-
- Flach KD, Zwart W. The first decade of estrogen receptor cistromics in breast cancer. J Endocrinol. 2016;229(2):R43–R56. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

