Novel peptides for deciphering structural and signalling functions of E-cadherin in mouse embryonic stem cells
- PMID: 28169326
- PMCID: PMC5294416
- DOI: 10.1038/srep41827
Novel peptides for deciphering structural and signalling functions of E-cadherin in mouse embryonic stem cells
Abstract
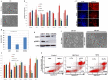
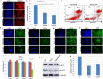
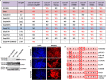
We have previously shown that E-cadherin regulates the naive pluripotent state of mouse embryonic stem cells (mESCs) by enabling LIF-dependent STAT3 phosphorylation, with E-cadherin null mESCs exhibiting over 3000 gene transcript alterations and a switch to Activin/Nodal-dependent pluripotency. However, elucidation of the exact mechanisms associated with E-cadherin function in mESCs is compounded by the difficulty in delineating the structural and signalling functions of this protein. Here we show that mESCs treated with the E-cadherin neutralising antibody DECMA-1 or the E-cadherin binding peptide H-SWELYYPLRANL-NH2 (Epep) exhibit discrete profiles for pluripotent transcripts and NANOG protein expression, demonstrating that the type of E-cadherin inhibitor employed dictates the cellular phenotype of mESCs. Alanine scanning mutation of Epep revealed residues critical for Tbx3, Klf4 and Esrrb transcript repression, cell-cell contact abrogation, cell survival in suspension, STAT3 phosphorylation and water solubility. STAT3 phosphorylation was found to be independent of loss of cell-cell contact and Activin/Nodal-dependent pluripotency and a peptide is described that enhances STAT3 phosphorylation and Nanog transcript and protein expression in mESCs. These peptides represent a useful resource for deciphering the structural and signalling functions of E-cadherin and demonstrate that complete absence of E-cadherin protein is likely required for hierarchical signalling pathway alterations in mESCs.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

