RNA-Seq analysis of nodule development at five different developmental stages of soybean (Glycine max) inoculated with Bradyrhizobium japonicum strain 113-2
- PMID: 28169364
- PMCID: PMC5294573
- DOI: 10.1038/srep42248
RNA-Seq analysis of nodule development at five different developmental stages of soybean (Glycine max) inoculated with Bradyrhizobium japonicum strain 113-2
Abstract
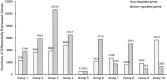
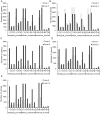
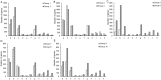
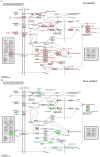
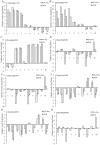
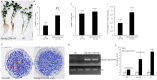
Nodule development directly affects nitrogen fixation efficiency during soybean growth. Although abundant genome-based information related to nodule development has been released and some studies have reported the molecular mechanisms that regulate nodule development, information on the way nodule genes operate in nodule development at different developmental stages of soybean is limited. In this report, notably different nodulation phenotypes in soybean roots inoculated with Bradyrhizobium japonicum strain 113-2 at five developmental stages (branching stage, flowering stage, fruiting stage, pod stage and harvest stage) were shown, and the expression of nodule genes at these five stages was assessed quantitatively using RNA-Seq. Ten comparisons were made between these developmental periods, and their differentially expressed genes were analysed. Some important genes were identified, primarily encoding symbiotic nitrogen fixation-related proteins, cysteine proteases, cystatins and cysteine-rich proteins, as well as proteins involving plant-pathogen interactions. There were no significant shifts in the distribution of most GO functional annotation terms and KEGG pathway enrichment terms between these five development stages. A cystatin Glyma18g12240 was firstly identified from our RNA-seq, and was likely to promote nodulation and delay nodule senescence. This study provides molecular material for further investigations into the mechanisms of nitrogen fixation at different soybean developmental stages.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

