Clustering of 770,000 genomes reveals post-colonial population structure of North America
- PMID: 28169989
- PMCID: PMC5309710
- DOI: 10.1038/ncomms14238
Clustering of 770,000 genomes reveals post-colonial population structure of North America
Abstract
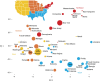
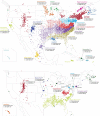
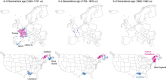
Despite strides in characterizing human history from genetic polymorphism data, progress in identifying genetic signatures of recent demography has been limited. Here we identify very recent fine-scale population structure in North America from a network of over 500 million genetic (identity-by-descent, IBD) connections among 770,000 genotyped individuals of US origin. We detect densely connected clusters within the network and annotate these clusters using a database of over 20 million genealogical records. Recent population patterns captured by IBD clustering include immigrants such as Scandinavians and French Canadians; groups with continental admixture such as Puerto Ricans; settlers such as the Amish and Appalachians who experienced geographic or cultural isolation; and broad historical trends, including reduced north-south gene flow. Our results yield a detailed historical portrait of North America after European settlement and support substantial genetic heterogeneity in the United States beyond that uncovered by previous studies.
Conflict of interest statement
The authors declare competing financial interests: authors affiliated with AncestryDNA may have equity in Ancestry; T.R. and S.S. were summer interns at Ancestry when they contributed to this work; E.B. received consultant fees from Ancestry; and a provisional patent application has been filed relating to this work (Application #15/168,011).
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

