Mouse Dux is myotoxic and shares partial functional homology with its human paralog DUX4
- PMID: 28173143
- PMCID: PMC5409219
- DOI: 10.1093/hmg/ddw287
Mouse Dux is myotoxic and shares partial functional homology with its human paralog DUX4
Abstract
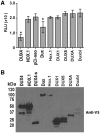
D4Z4 repeats are present in at least 11 different mammalian species, including humans and mice. Each repeat contains an open reading frame encoding a double homeodomain (DUX) family transcription factor. Aberrant expression of the D4Z4 ORF called DUX4 is associated with the pathogenesis of Facioscapulohumeral muscular dystrophy (FSHD). DUX4 is toxic to numerous cell types of different species, and over-expression caused dysmorphism and developmental arrest in frogs and zebrafish, embryonic lethality in transgenic mice, and lesions in mouse muscle. Because DUX4 is a primate-specific gene, questions have been raised about the biological relevance of over-expressing it in non-primate models, as DUX4 toxicity could be related to non-specific cellular stress induced by over-expressing a DUX family transcription factor in organisms that did not co-evolve its regulated transcriptional networks. We assessed toxic phenotypes of DUX family genes, including DUX4, DUX1, DUX5, DUXA, DUX4-s, Dux-bl and mouse Dux. We found that DUX proteins were not universally toxic, and only the mouse Dux gene caused similar toxic phenotypes as human DUX4. Using RNA-seq, we found that 80% of genes upregulated by Dux were similarly increased in DUX4-expressing cells. Moreover, 43% of Dux-responsive genes contained ChIP-seq binding sites for both Dux and DUX4, and both proteins had similar consensus binding site sequences. These results suggested DUX4 and Dux may regulate some common pathways, and despite diverging from a common progenitor under different selective pressures for millions of years, the two genes maintain partial functional homology.
Figures




References
-
- INSERM, French Ministry of Health. (2008), In Orphanet Report. Orphanet, Vol. 2008.
-
- Flanigan K.M., Harper S.Q. (2013) John Wiley & Sons, L; (ed.), In Muscle Disease, in press., pp. 288–297.
-
- Tawil R., Van Der Maarel S.M. (2006) Facioscapulohumeral muscular dystrophy. Muscle & Nerve, 34, 1–15. - PubMed
-
- Wijmenga C., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R., et al. (1992) Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet., 2, 26–30. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

