An approach of identifying differential nucleosome regions in multiple samples
- PMID: 28173752
- PMCID: PMC5297132
- DOI: 10.1186/s12864-017-3541-9
An approach of identifying differential nucleosome regions in multiple samples
Abstract
Background: Nucleosome plays a role in transcriptional regulation through occluding the binding of proteins to DNA sites. Nucleosome occupancy varies among different cell types. Identification of such variation will help to understand regulation mechanism. The previous researches focused on the methods for two-sample comparison. However, a multiple-sample comparison (n ≥ 3) is necessary, especially in studying development and cancer. METHODS: Here, we proposed a Chi-squared test-based approach, named as Dimnp, to identify differential nucleosome regions (DNRs) in multiple samples. Dimnp is designed for sequenced reads data and includes the modules of both calling nucleosome occupancy and identifying DNRs.
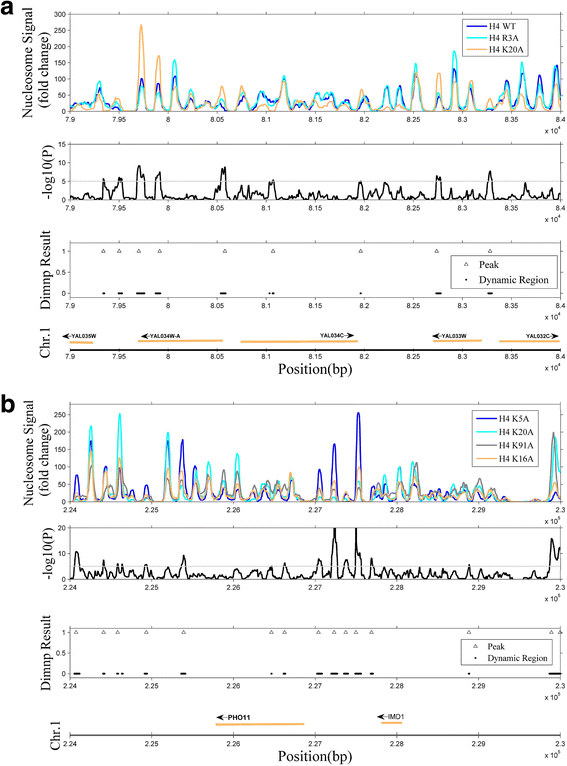
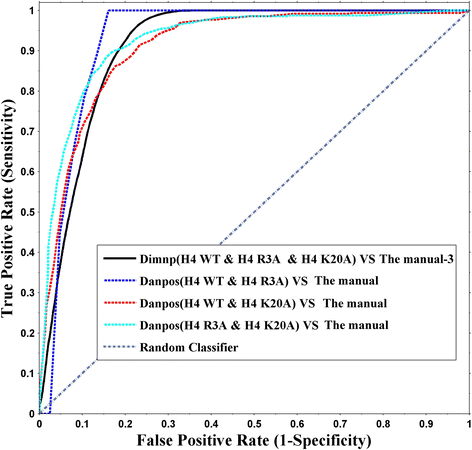
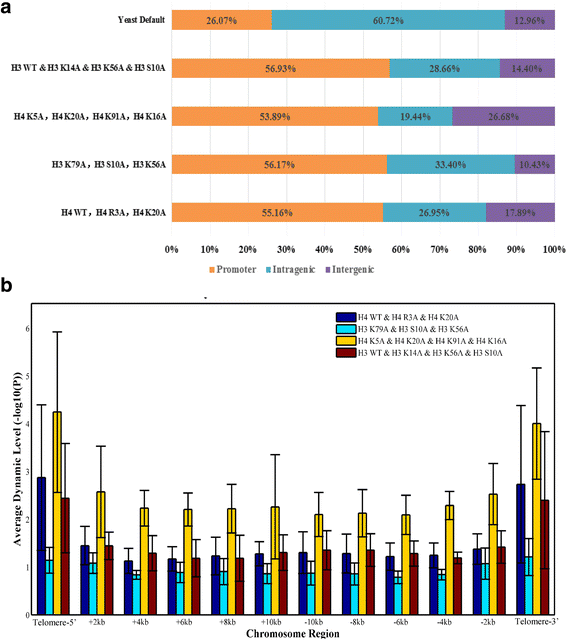
Results: We validated Dimnp on dataset of the mutant strains in which the modifiable histone residues are mutated into alanine in Saccharomyces cerevisiae. Dimnp shows a good capacity (area under the curve > 0.87) compared with the manually identified DNRs. Just by one time, Dimnp is able to identify all the DNRs identified by two-sample method Danpos. Under a deviation of 40 bp, the matched DNRs are above 60% between Dimnp and Danpos. With Dimnp, we found that promoters and telomeres are highly dynamic upon mutating the modifiable histone residues.
Conclusions: We developed a tool of identifying the DNRs in multiple samples and cell types. The tool can be applied in studying nucleosome variation in gradual change in development and cancer.
Keywords: Chi-squared test; Differential nucleosome regions (DNRs); Multiple cell types; Nucleosome.
Figures


Similar articles
-
DNMHMM: An approach to identify the differential nucleosome regions in multiple cell types based on a Hidden Markov Model.Biosystems. 2019 Nov;185:104033. doi: 10.1016/j.biosystems.2019.104033. Epub 2019 Sep 18. Biosystems. 2019. PMID: 31541672
-
DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing.Genome Res. 2013 Feb;23(2):341-51. doi: 10.1101/gr.142067.112. Epub 2012 Nov 28. Genome Res. 2013. PMID: 23193179 Free PMC article.
-
A deformation energy-based model for predicting nucleosome dyads and occupancy.Sci Rep. 2016 Apr 7;6:24133. doi: 10.1038/srep24133. Sci Rep. 2016. PMID: 27053067 Free PMC article.
-
Prediction of nucleosome occupancy in Saccharomyces cerevisiae using position-correlation scoring function.Genomics. 2011 Nov;98(5):359-66. doi: 10.1016/j.ygeno.2011.07.008. Epub 2011 Aug 2. Genomics. 2011. PMID: 21839161
-
Genome-wide analysis predicts DNA structural motifs as nucleosome exclusion signals.Mol Biosyst. 2009 Dec;5(12):1703-12. doi: 10.1039/b905132e. Epub 2009 May 29. Mol Biosyst. 2009. PMID: 19587895
Cited by
-
Application of Next-Generation Sequencing in Neurodegenerative Diseases: Opportunities and Challenges.Neuromolecular Med. 2021 Jun;23(2):225-235. doi: 10.1007/s12017-020-08601-7. Epub 2020 May 12. Neuromolecular Med. 2021. PMID: 32399804 Review.
-
NucMap: a database of genome-wide nucleosome positioning map across species.Nucleic Acids Res. 2019 Jan 8;47(D1):D163-D169. doi: 10.1093/nar/gky980. Nucleic Acids Res. 2019. PMID: 30335176 Free PMC article.
-
Nucleosome Dynamics: a new tool for the dynamic analysis of nucleosome positioning.Nucleic Acids Res. 2019 Oct 10;47(18):9511-9523. doi: 10.1093/nar/gkz759. Nucleic Acids Res. 2019. PMID: 31504766 Free PMC article.
-
Mapping nucleosome and chromatin architectures: A survey of computational methods.Comput Struct Biotechnol J. 2022 Jul 26;20:3955-3962. doi: 10.1016/j.csbj.2022.07.037. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35950186 Free PMC article. Review.
References
-
- Riffo-Campos AL, Castillo J, Tur G, Gonzalez-Figueroa P, Georgieva EI, Rodriguez JL, Lopez-Rodas G, Rodrigo MI, Franco L. Nucleosome-specific, time-dependent changes in histone modifications during activation of the early growth response 1 (Egr1) gene. J Biol Chem. 2015;290(1):197–208. doi: 10.1074/jbc.M114.579292. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

