Identification of influenza A nucleoprotein body domain residues essential for viral RNA expression expose antiviral target
- PMID: 28173821
- PMCID: PMC5294902
- DOI: 10.1186/s12985-017-0694-8
Identification of influenza A nucleoprotein body domain residues essential for viral RNA expression expose antiviral target
Abstract
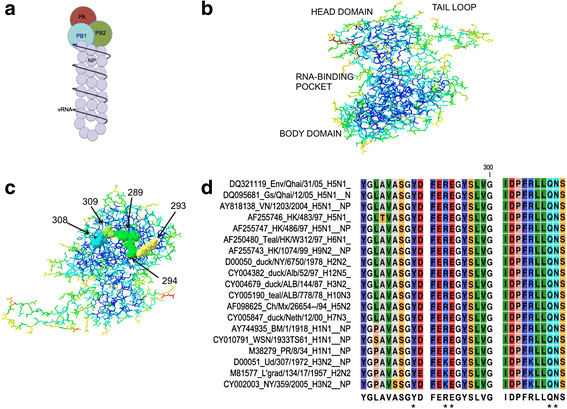
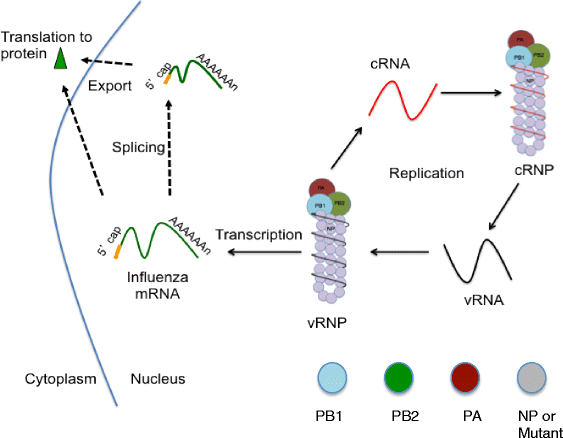
Background: Influenza A virus is controlled with yearly vaccination while emerging global pandemics are kept at bay with antiviral medications. Unfortunately, influenza A viruses have emerged resistance to approved influenza antivirals. Accordingly, there is an urgent need for novel antivirals to combat emerging influenza A viruses resistant to current treatments. Conserved viral proteins are ideal targets because conserved protein domains are present in most, if not all, influenza subtypes, and are presumed less prone to evolve viable resistant versions. The threat of an antiviral resistant influenza pandemic justifies our study to identify and characterize antiviral targets within influenza proteins that are highly conserved. Influenza A nucleoprotein (NP) is highly conserved and plays essential roles throughout the viral lifecycle, including viral RNA synthesis.
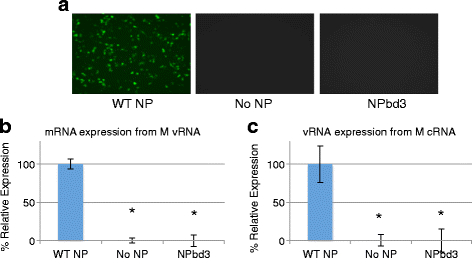
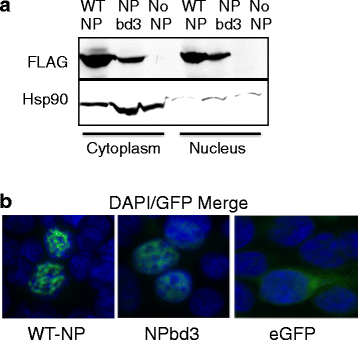
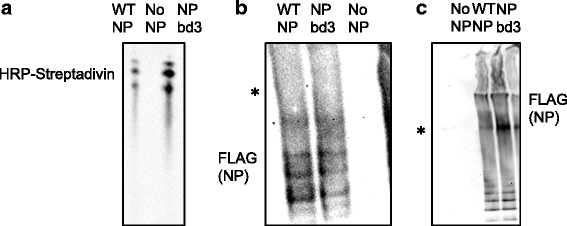
Methods: Using NP crystal structure, we targeted accessible amino acids for substitution. To characterize the NP proteins, reconstituted viral ribonucleoproteins (vRNPs) were expressed in 293 T cells, RNA was isolated, and reverse transcription - quantitative PCR (RT-qPCR) was employed to assess viral RNA expressed from reconstituted vRNPs. Location was confirmed using cellular fractionation and western blot, along with observation of NP-GFP fusion proteins. Nucleic acid binding, oligomerization, and vRNP formation, were each assessed with native gel electrophoresis.
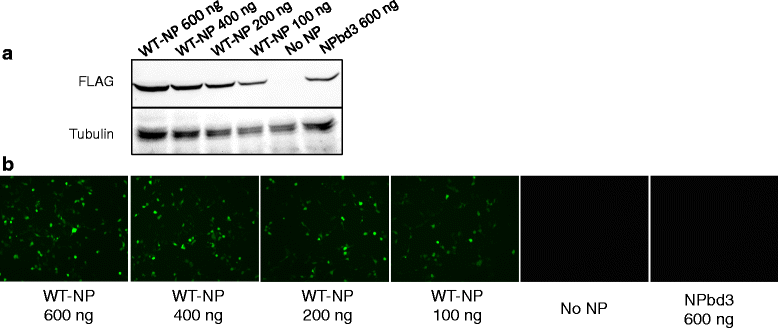
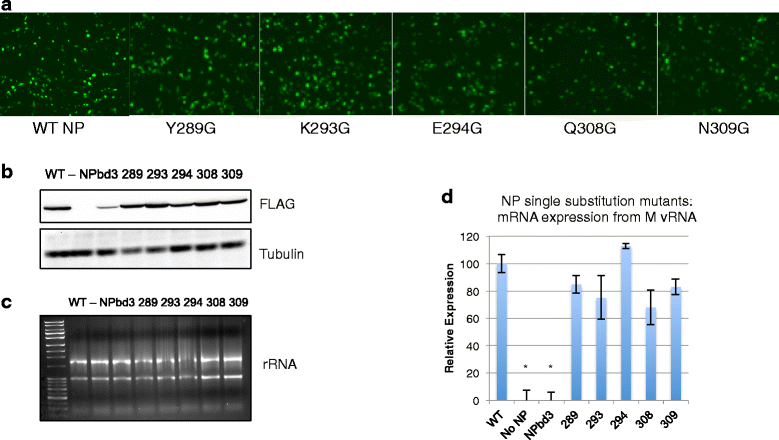
Results: Here we report characterization of an accessible and conserved five amino acid region within the NP body domain that plays a redundant but essential role in viral RNA synthesis. Our data demonstrate substitutions in this domain did not alter NP localization, oligomerization, or ability to bind nucleic acids, yet resulted in a defect in viral RNA expression. To define this region further, single and double amino acid substitutions were constructed and investigated. All NP single substitutions were functional, suggesting redundancy, yet different combinations of two amino acid substitutions resulted in a significant defect in RNA expression, confirming these accessible amino acids in the NP body domain play an important role in viral RNA synthesis.
Conclusions: The identified conserved and accessible NP body domain represents a viable antiviral target to counter influenza replication and this research will contribute to the well-informed design of novel therapies to combat emerging influenza viruses.
Keywords: Influenza; Nucleoprotein; RNA; Virus.
Figures
Similar articles
-
A Novel Antiviral Target Structure Involved in the RNA Binding, Dimerization, and Nuclear Export Functions of the Influenza A Virus Nucleoprotein.PLoS Pathog. 2015 Jul 29;11(7):e1005062. doi: 10.1371/journal.ppat.1005062. eCollection 2015 Jul. PLoS Pathog. 2015. PMID: 26222066 Free PMC article.
-
Nuclear localized Influenza nucleoprotein N-terminal deletion mutant is deficient in functional vRNP formation.Virol J. 2014 Aug 31;11:155. doi: 10.1186/1743-422X-11-155. Virol J. 2014. PMID: 25174360 Free PMC article.
-
Using mutagenesis to explore conserved residues in the RNA-binding groove of influenza A virus nucleoprotein for antiviral drug development.Sci Rep. 2016 Feb 26;6:21662. doi: 10.1038/srep21662. Sci Rep. 2016. PMID: 26916998 Free PMC article.
-
Influenza virus nucleoprotein: structure, RNA binding, oligomerization and antiviral drug target.Future Microbiol. 2013 Dec;8(12):1537-45. doi: 10.2217/fmb.13.128. Future Microbiol. 2013. PMID: 24266354 Review.
-
Structure and sequence analysis of influenza A virus nucleoprotein.Sci China C Life Sci. 2009 May;52(5):439-49. doi: 10.1007/s11427-009-0064-x. Epub 2009 May 27. Sci China C Life Sci. 2009. PMID: 19471866 Review.
Cited by
-
An overview of influenza A virus genes, protein functions, and replication cycle highlighting important updates.Virus Genes. 2022 Aug;58(4):255-269. doi: 10.1007/s11262-022-01904-w. Epub 2022 Apr 26. Virus Genes. 2022. PMID: 35471490 Review.
-
A Review and Meta-Analysis of Influenza Interactome Studies.Front Microbiol. 2022 Apr 21;13:869406. doi: 10.3389/fmicb.2022.869406. eCollection 2022. Front Microbiol. 2022. PMID: 35531276 Free PMC article. Review.
-
Gene Segment Interactions Can Drive the Emergence of Dominant Yet Suboptimal Gene Constellations During Influenza Virus Reassortment.Front Microbiol. 2021 Jul 14;12:683152. doi: 10.3389/fmicb.2021.683152. eCollection 2021. Front Microbiol. 2021. PMID: 34335507 Free PMC article.
-
Sudan Ebolavirus VP35-NP Crystal Structure Reveals a Potential Target for Pan-Filovirus Treatment.mBio. 2019 Jul 23;10(4):e00734-19. doi: 10.1128/mBio.00734-19. mBio. 2019. PMID: 31337716 Free PMC article.
-
BIK polymorphism and proteasome regulation unveil host risk factor for severe influenza.Proc Natl Acad Sci U S A. 2025 Jul 15;122(28):e2424367122. doi: 10.1073/pnas.2424367122. Epub 2025 Jul 8. Proc Natl Acad Sci U S A. 2025. PMID: 40627391 Free PMC article.
References
-
- Centers for Disease Control and Prevention. http://www.cdc.gov/flu/avianflu/avian-in-humans.htm. Accessed 15 June 2016.
-
- Centers for Disease Control and Prevention. http://www.cdc.gov/flu/about/viruses/transmission.htm. Accessed 15 June 2016.
-
- Centers for Disease Control and Prevention. http://www.cdc.gov/flu/professionals/vaccination/effectiveness-studies.htm. Accessed 15 June 2016.
-
- Centers for Disease Control and Prevention. http://www.cdc.gov/media/releases/2016/s0622-laiv-flu.html. Accessed 15 July 2016.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

