Microbiomic differences in tumor and paired-normal tissue in head and neck squamous cell carcinomas
- PMID: 28173873
- PMCID: PMC5297129
- DOI: 10.1186/s13073-017-0405-5
Microbiomic differences in tumor and paired-normal tissue in head and neck squamous cell carcinomas
Abstract
Background: While the role of the gut microbiome in inflammation and colorectal cancers has received much recent attention, there are few data to support an association between the oral microbiome and head and neck squamous cell carcinomas. Prior investigations have been limited to comparisons of microbiota obtained from surface swabs of the oral cavity. This study aims to identify microbiomic differences in paired tumor and non-tumor tissue samples in a large group of 121 patients with head and neck squamous cell carcinomas and correlate these differences with clinical-pathologic features.
Methods: Total DNA was extracted from paired normal and tumor resection specimens from 169 patients; 242 samples from 121 patients were included in the final analysis. Microbiomic content of each sample was determined using 16S rDNA amplicon sequencing. Bioinformatic analysis was performed using QIIME algorithms. F-testing on cluster strength, Wilcoxon signed-rank testing on differential relative abundances of paired tumor-normal samples, and Wilcoxon rank-sum testing on the association of T-stage with relative abundances were conducted in R.
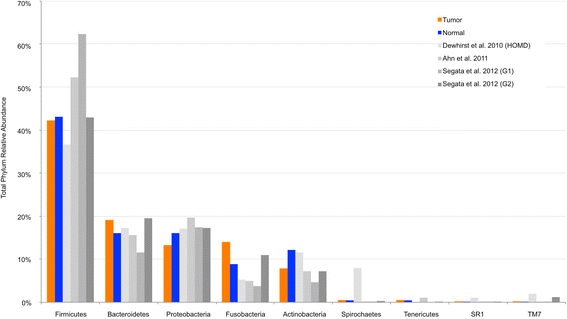
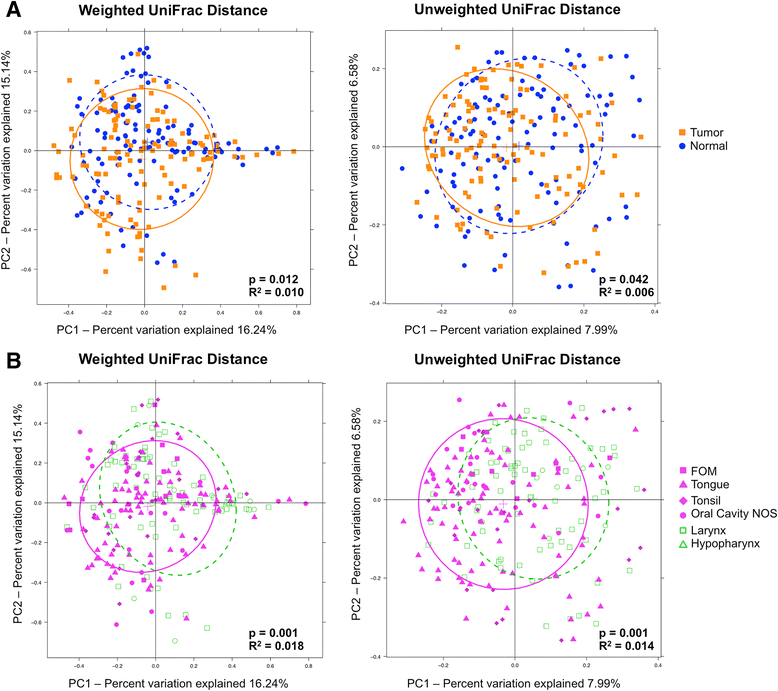
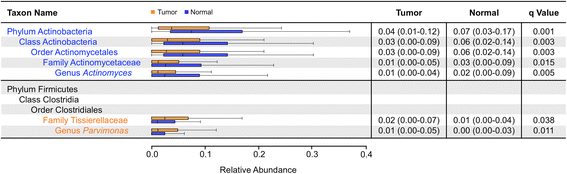
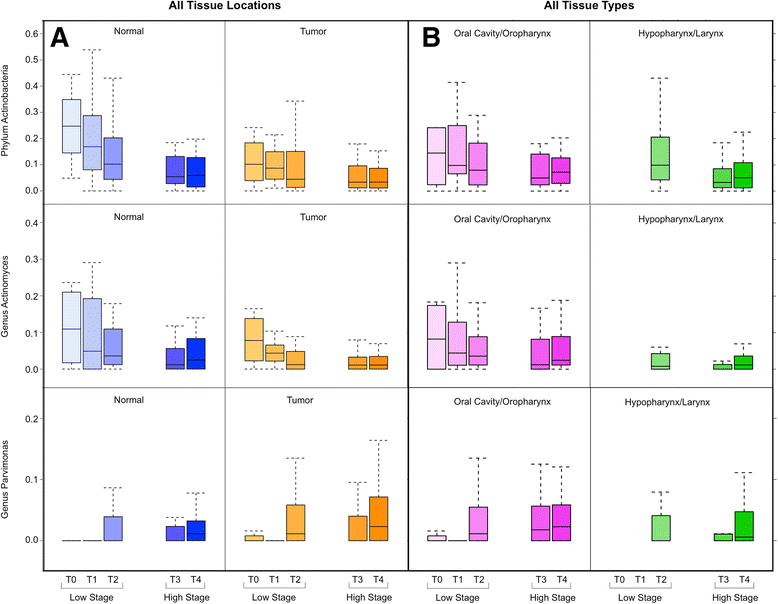
Results: We observed no significant difference in measures of alpha diversity between tumor and normal tissue (Shannon index: p = 0.13, phylogenetic diversity: p = 0.42). Similarly, although we observed statistically significantly differences in both weighted (p = 0.01) and unweighted (p = 0.04) Unifrac distances between tissue types, the tumor/normal grouping explained only a small proportion of the overall variation in the samples (weighted R2 = 0.01, unweighted R2 < 0.01). Notably, however, when comparing the relative abundances of individual taxa between matched pairs of tumor and normal tissue, we observed that Actinomyces and its parent taxa up to the phylum level were significantly depleted in tumor relative to normal tissue (q < 0.01), while Parvimonas was increased in tumor relative to normal tissue (q = 0.01). These differences were more pronounced among patients with more extensive disease as measured by higher T-stage.
Conclusions: Matched pairs analysis of individual tumor-normal pairs revealed significant differences in relative abundance of specific taxa, namely in the genus Actinomyces. These differences were more pronounced among patients with higher T-stage. Our observations suggest further experiments to interrogate potential novel mechanisms relevant to carcinogenesis associated with alterations of the oral microbiome that may have consequences for the human host.
Keywords: Bacteria; Head and neck squamous cell carcinoma (HNSCC); Microbiome.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

