L-OPA1 regulates mitoflash biogenesis independently from membrane fusion
- PMID: 28174208
- PMCID: PMC5331265
- DOI: 10.15252/embr.201642931
L-OPA1 regulates mitoflash biogenesis independently from membrane fusion
Abstract
Mitochondrial flashes mediated by optic atrophy 1 (OPA1) fusion protein are bioenergetic responses to stochastic drops in mitochondrial membrane potential (Δψm) whose origin is unclear. Using structurally distinct genetically encoded pH-sensitive probes, we confirm that flashes are matrix alkalinization transients, thereby establishing the pH nature of these events, which we renamed "mitopHlashes". Probes located in cristae or intermembrane space as verified by electron microscopy do not report pH changes during Δψm drops or respiratory chain inhibition. Opa1 ablation does not alter Δψm fluctuations but drastically decreases the efficiency of mitopHlash/Δψm coupling, which is restored by re-expressing fusion-deficient OPA1K301A and preserved in cells lacking the outer-membrane fusion proteins MFN1/2 or the OPA1 proteases OMA1 and YME1L, indicating that mitochondrial membrane fusion and OPA1 proteolytic processing are dispensable. pH/Δψm uncoupling occurs early during staurosporine-induced apoptosis and is mitigated by OPA1 overexpression, suggesting that OPA1 maintains mitopHlash competence during stress conditions. We propose that OPA1 stabilizes respiratory chain supercomplexes in a conformation that enables respiring mitochondria to compensate a drop in Δψm by an explosive matrix pH flash.
Keywords: OPA1; bioenergetics; membrane fusion; mitoflash.
© 2017 The Authors. Published under the terms of the CC BY 4.0 license.
Figures
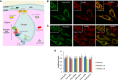
- A
Predicted mitochondrial localization of pH sensors used in this study. pH‐sensitive fluorescent proteins were addressed to the mitochondrial matrix, the intermembrane space (IMS), or the cristae. MPP‐spHluorin, mito‐sypHer, and Cox8‐pHred are expressed in the matrix. Smac‐rpHluorin is targeted to the IMS. CIV8‐spHluorin is fused to the complex IV subunit 8a and faces the intra‐cristae space, while CVe‐ and CVɣ‐spHluorins are fused to the complex V and are located on the cristae side and on the matrix side, respectively. Complexes formed by long (L‐OPA1) and short forms (S‐OPA1) of OPA1 are located at the cristae junctions. OMM: outer mitochondrial membrane, IMM: inner mitochondrial membrane, CI: complex I, CII: complex II, CIII: complex III, CIV: complex IV, CV: complex V.
- B–D
Colocalization between the endogenous mitochondrial marker Hsp60 and the different pH sensors. Images of HeLa cells showing endogenous Hsp60 (red signal) and (B) Smac‐rpHluorin or (C) CVɣ‐spHluorin (green signal) and their overlays. The insets show higher magnification of the regions outlined by yellow rectangles. Scale bars: 10 μm. (D) Analysis of colocalization is represented by Pearson's coefficient (indicating the correlation between Hsp60 and the pH probe signals) and by the Mander's A (representing the proportion of Hsp60 signal overlapping with the pH probes) and Mander's B coefficients (representing the proportion of the pH probe signals overlapping with Hsp60). Data are means ± SD of three independent experiments.

- A–C
Confocal images of (A) Cox8‐pHred, (B) CVɣ‐spHluorin, and (C) MPP‐spHluorin in HeLa cells. Scale bars: 10 μm.
- D
In situ pH calibration of pHred (F 561/F 405 excitation ratio), CVɣ‐, and MPP‐spHluorins (λex: 488 nm).
- E
Average resting pH reported by the three probes. Box and whisker plots show the 25th–75th percentiles, median, and maximal values from three independent experiments.
- F
Representative curve showing change in Cox8‐pHred F 561/F 405 ratio upon addition of antimycin A (AntA, 5 μM), corresponding to an acidification of the matrix. Similar responses were obtained with CVɣ‐ and MPP‐spHluorins (Fig EV3A and B).
- G
Antiparallel changes in Cox8‐pHred fluorescence at λex of 405 and 561 nm corresponding to matrix alkalinization transients.
- H, I
Simultaneous recordings of TMRM fluorescence and matrix pH with (H) CVɣ‐ or (I) MPP‐spHluorin reporting alkalinization events during drops in Δψm.
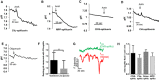
- A–D
Effect of antimycin in HeLa cells expressing spHluorins targeted to matrix or cristae. Change in fluorescence intensity of (A) CVɣ‐, (B) MPP‐, (C) CIV8‐, and (D) CVe‐spHluorins (λex: 488 nm) evoked by the addition of antimycin A (AntA). Fluorescence decreases correspond to an acidification of the mitochondrial compartment.
- E
Effect of oligomycin in HeLa cells expressing Smac‐rpHluorin.
- F
Effect of galactose and low glucose media on resting pH values measured with CVe‐spHluorin (n = 20 and 19 cells for each medium, respectively). Values are mean ± SD of three independent experiments. Unpaired t‐test with Welch's correction, **P = 0.0023.
- G
CIV8‐spHluorin recordings showing the absence of pH transients during drops in Δψm in cells cultured with galactose. Identical results were obtained with CVe‐spHluorin and Smac‐rpHluorin.
- H
MitopHlash frequency in HeLa cells co‐expressing mito‐sypHer and Smac‐rpHluorin or spHluorins fused to CIV8, CVe, or MPP. mito‐sypHer flashing activity persisted unabatedly in cells expressing pH sensors in the intra‐cristae space or in the IMS. **P < 0.01.
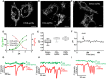
- A–C
Confocal images of (A) CIV8‐spHluorin, (B) CVe‐spHluorin, and (C) Smac‐rpHluorin in HeLa cells. Scale bars: 10 μm.
- D
In situ pH calibration of Smac‐rpHluorin (F 488/F 405 excitation ratio), CIV8‐ and CVe‐spHluorins (λex: 488 nm).
- E
Average resting pH reported by the three probes. Box and whisker plots show the 25th–75th percentiles, median, and maximal values from three independent experiments.
- F
Representative curve showing change in Smac‐rpHluorin fluorescence evoked by 5 μM antimycin A (AntA). Similar responses were obtained with CIV8‐ and CVe‐spHluorins (Fig EV3C and D).
- G–I
Concomitant recordings of Δψm (TMRM) and pH measured with (G) CIV8‐, (H) CVe‐spHluorin, or (I) Smac‐rpHluorin reporting no detectable pH changes during drops in Δψm.
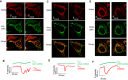
- A–F
Confocal images of cells expressing (A) CVe‐spHluorin, (C) Smac‐rpHluorin, or (E) CIV8‐spHluorin (green signals) loaded with TMRM (red), and time‐resolved recordings of changes in TMRM and (B) CVe‐spHluorin, (D) Smac‐rpHluorin, or (F) CIV8‐spHluorin fluorescence. The mitochondrial area of depolarization is greatly enhanced in cells expressing DRP1K38A, but pH changes remain undetectable with the pHluorins. Scale bars: 10 μm.
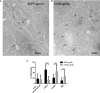
- A, B
Electron microscopy images showing the localization of gold‐labeled anti‐GFP antibodies in HeLa cells expressing (A) MPP‐ and (B) CIV8‐spHluorins. The IMM and OMM of mitochondria are outlined in gray to better visualize the different mitochondrial sub‐compartments. Black opened arrows, gray filled arrows, and blue arrowheads correspond to a cristae, matrix, and IMS localization, respectively. Scale bars: 100 nm.
- C
Distribution of gold particles in the different compartments. Values are means ± SD. 81 mitochondria with 269 gold particles were analyzed for MPP‐spHluorin and 47 mitochondria with 169 gold particles for CIV8‐spHluorin. Two‐way ANOVA, *P = 0.0211, ***P = 0.0005, ****P < 0.0001.
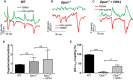
- A–C
Concurrent recordings of Δψm (TMRM) and matrix pH (mito‐sypHer, λex: 488 nm) in (A) WT, (B) Opa1 −/−, or (C) Opa1 −/− cells re‐expressing OPA1.
- D, E
Depolarization frequency (D) with n = 41, 25, and 42 cells recorded in WT, Opa1 −/−, and Opa1 −/− cells re‐expressing OPA1, respectively, and coupling between mitopHlashes and Δψm (E) (number of mitopHlashes/number of depolarizations) in the indicated cells with n = 65, 59, and 84 depolarization events recorded in 41, 25, and 42 WT, Opa1 −/− and Opa1 −/− cells re‐expressing OPA1, respectively. Values are means ± SD of three independent experiments. One‐way ANOVA with multiple comparisons, **P = 0.0013, ****P < 0.0001, ns: not significant.
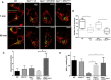
- A
Confocal images of matrix‐targeted photoactivable GFP (mito‐PA‐GFP) and mito‐DsRed co‐expressed in WT, Opa1 −/− cells transfected with control plasmid (ctr, pcDNA3), OPA1, or OPA1K301A. Mitochondrial fusion was assessed by tracking the area of PA‐GFP fluorescence 1 min (top) and 60 min (bottom) after photoactivation in the same cell. Scale bars: 10 μm.
- B
Quantification of mitochondrial fusion as percentage of PA‐GFP fluorescent area increase. Box and whisker plots show the 25th–75th percentiles, median, and maximal values from three independent experiments (n = 7, 6, 6, and 7 cells for WT, Opa1 −/− cells transfected with control plasmid (ctr, pcDNA3), OPA1, or OPA1K301A, respectively). One‐way ANOVA with multiple comparisons, *P = 0.0463 (WT vs. Opa1 −/− + ctr), *P = 0.0329 (Opa1 −/− + WT OPA1 vs. Opa1 −/− + OPA1 K301A).
- C, D
Depolarization frequency (C) and mitopHlash/Δψm coupling (D) in WT, Mfn1/2 −/−, and Opa1 −/− cells and in Opa1 −/− cells re‐expressing OPA1 or the fusion‐deficient mutant OPA1K301A. Values are means ± SD of three independent experiments with n = 129, 79, 194, 167, and 177 depolarization events recorded in 30, 28, 54, 43, and 41 cells, respectively. One‐way ANOVA with multiple comparisons, *P = 0.0407 (Opa1 −/− + ctr vs. Opa1 −/− + OPA1), *P = 0.0172 (Opa1 −/− + ctr vs. Opa1 −/− + OPA1K301A), **P = 0.0019.
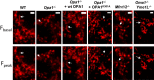
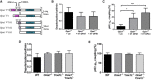
- A
Schematic representation of the long and short forms of OPA1 derived from splice variants 1 (V1) and 7 (V7) and their corresponding short forms produced by proteolytic cleavage at sites S1 or S2 by OMA1 or YME1L, respectively.
- B, C
Depolarization frequency (B) and mitopHlash/Δψm coupling (C) in Opa1 −/− cells encoding pcDNA3 (ctr), re‐expressing the long V1 or V7 OPA1 variant. n = 102, 198, and 93 depolarization events recorded in 21, 69, and 29 cells, respectively.
- D, E
Depolarization frequency (D) and mitopHlash/Δψm coupling (E) in WT, Oma1 −/−, Yme1L −/−, or Oma1 −/− Yme1L −/− cells. n = 94, 100, 77, and 90 depolarization events recorded in 41, 40, 39, and 43 cells, respectively.
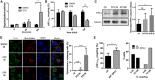
- A, B
Depolarization frequency (A) and mitopHlash/Δψm coupling (B) in WT cells exposed to DMSO or 1 μM STS. Values are means ± SD of three independent experiments. Two‐way ANOVA, *P = 0.0163, **P = 0.0085. n = 72, 99, 58 depolarization events recorded in 22, 27, and 20 cells for DMSO and 62, 75, 103 depolarization events recorded in 18, 22, and 24 cells for STS at 0, 60, and 120 min, respectively.
- C
Western blots of OPA1 and tubulin performed on cytosolic fractions from WT cells exposed to DMSO or STS. The quantification of cytosolic OPA1 normalized to the amount of tubulin for each condition is shown. Values are means ± SD of three independent experiments, *P = 0.0332, paired t‐test.
- D
Confocal images showing cytochrome c immunoreactivity (green), mito‐DsRed (red), and DAPI (blue) fluorescence 2 or 6 h after STS exposure. Note the mitochondrial localization of cytochrome c after 2 h of STS. Scale bars: 10 μm. Right panel shows the fraction of cells displaying cytoplasmic cytochrome c immunoreactivity. Values are means ± SD of three independent experiments, ****P < 0.0001, one‐way ANOVA with multiple comparisons.
- E
mitopHlash/Δψm coupling 2 h after STS exposure in WT MEF cells overexpressing OPA1 (variant 1) or control plasmid (ctr). Values are means ± SD of three independent experiments. Unpaired t‐test with Welch's correction, *P = 0.0293. The fraction of cells with low pH/Δψm coupling (0–25%) and high pH/Δψm coupling (75–100%) is indicated. n = 422 and 312 depolarization events recorded in 57 and 44 cells for control (ctr) and OPA1 re‐expression.
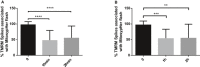
- A, B
mitopHlash/Δψm coupling in WT MEF cells loaded with TMRM and exposed to (A) 250 mM H2O2 for 10 and 20 min or (B) 100 μM etoposide for 1 or 2 h. n = 38, 97, 98 depolarization events recorded in 25, 18, and 22 cells at 0, 10, and 20 min of H2O2 treatment, respectively, and n = 27, 111, 34 depolarization events recorded in 16, 18, and 13 cells at 0, 1, and 2 h of etoposide treatment, respectively. Values are means ± SD of three independent experiments. One‐way ANOVA with multiple comparisons, **P = 0.0018, ***P = 0.0005, ****P < 0.0001.
References
-
- Hatefi Y (1985) The mitochondrial electron transport and oxidative phosphorylation system. Annu Rev Biochem 54: 1015–1069 - PubMed
-
- Mitchell P (1961) Coupling of phosphorylation to electron and hydrogen transfer by a chemi‐osmotic type of mechanism. Nature 191: 144–148 - PubMed
-
- Mitchell P (1966) Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Biol Rev Camb Philos Soc 41: 445–502 - PubMed
-
- Nicholls DG (1974) The influence of respiration and ATP hydrolysis on the proton‐electrochemical gradient across the inner membrane of rat‐liver mitochondria as determined by ion distribution. Eur J Biochem 50: 305–315 - PubMed
-
- Rottenberg H (1975) The measurement of transmembrane electrochemical proton gradients. J Bioenerg 7: 61–74 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

