Respiratory Complex I in Bos taurus and Paracoccus denitrificans Pumps Four Protons across the Membrane for Every NADH Oxidized
- PMID: 28174301
- PMCID: PMC5377811
- DOI: 10.1074/jbc.M116.771899
Respiratory Complex I in Bos taurus and Paracoccus denitrificans Pumps Four Protons across the Membrane for Every NADH Oxidized
Abstract
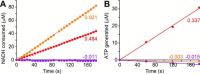
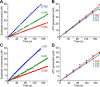
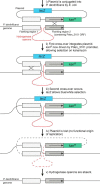
Respiratory complex I couples electron transfer between NADH and ubiquinone to proton translocation across an energy-transducing membrane to support the proton-motive force that drives ATP synthesis. The proton-pumping stoichiometry of complex I (i.e. the number of protons pumped for each two electrons transferred) underpins all mechanistic proposals. However, it remains controversial and has not been determined for any of the bacterial enzymes that are exploited as model systems for the mammalian enzyme. Here, we describe a simple method for determining the proton-pumping stoichiometry of complex I in inverted membrane vesicles under steady-state ADP-phosphorylating conditions. Our method exploits the rate of ATP synthesis, driven by oxidation of NADH or succinate with different sections of the respiratory chain engaged in catalysis as a proxy for the rate of proton translocation and determines the stoichiometry of complex I by reference to the known stoichiometries of complexes III and IV. Using vesicles prepared from mammalian mitochondria (from Bos taurus) and from the bacterium Paracoccus denitrificans, we show that four protons are pumped for every two electrons transferred in both cases. By confirming the four-proton stoichiometry for mammalian complex I and, for the first time, demonstrating the same value for a bacterial complex, we establish the utility of P. denitrificans complex I as a model system for the mammalian enzyme. P. denitrificans is the first system described in which mutagenesis in any complex I core subunit may be combined with quantitative proton-pumping measurements for mechanistic studies.
Keywords: complex I; electron transfer complex; mitochondria; mitochondrial respiratory chain complex; oxidative phosphorylation; proton motive force.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

References
-
- Hirst J. (2013) Mitochondrial complex I. Annu. Rev. Biochem. 82, 551–575 - PubMed
-
- Hirst J., Carroll J., Fearnley I. M., Shannon R. J., and Walker J. E. (2003) The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim. Biophys. Acta 1604, 135–150 - PubMed
-
- Walker J. E. (1992) The NADH:ubiquinone oxidoreductase (complex I) of respiratory chains. Q. Rev. Biophys. 25, 253–324 - PubMed
-
- Zickermann V., Wirth C., Nasiri H., Siegmund K., Schwalbe H., Hunte C., and Brandt U. (2015) Mechanistic insight from the crystal structure of mitochondrial complex I. Science 347, 44–49 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

