A Mobile Element in mutS Drives Hypermutation in a Marine Vibrio
- PMID: 28174306
- PMCID: PMC5296598
- DOI: 10.1128/mBio.02045-16
A Mobile Element in mutS Drives Hypermutation in a Marine Vibrio
Abstract
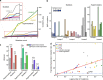
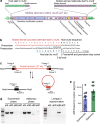
Bacteria face a trade-off between genetic fidelity, which reduces deleterious mistakes in the genome, and genetic innovation, which allows organisms to adapt. Evidence suggests that many bacteria balance this trade-off by modulating their mutation rates, but few mechanisms have been described for such modulation. Following experimental evolution and whole-genome resequencing of the marine bacterium Vibrio splendidus 12B01, we discovered one such mechanism, which allows this bacterium to switch to an elevated mutation rate. This switch is driven by the excision of a mobile element residing in mutS, which encodes a DNA mismatch repair protein. When integrated within the bacterial genome, the mobile element provides independent promoter and translation start sequences for mutS-different from the bacterium's original mutS promoter region-which allow the bacterium to make a functional mutS gene product. Excision of this mobile element rejoins the mutS gene with host promoter and translation start sequences but leaves a 2-bp deletion in the mutS sequence, resulting in a frameshift and a hypermutator phenotype. We further identified hundreds of clinical and environmental bacteria across Betaproteobacteria and Gammaproteobacteria that possess putative mobile elements within the same amino acid motif in mutS In a subset of these bacteria, we detected excision of the element but not a frameshift mutation; the mobile elements leave an intact mutS coding sequence after excision. Our findings reveal a novel mechanism by which one bacterium alters its mutation rate and hint at a possible evolutionary role for mobile elements within mutS in other bacteria.
Importance: DNA mutations are a double-edged sword. Most mutations are harmful; they can scramble precise genetic sequences honed over thousands of generations. However, in rare cases, mutations also produce beneficial new traits that allow populations to adapt to changing environments. Recent evidence suggests that some bacteria balance this trade-off by altering their mutation rates to suit their environment. To date, however, we know of few mechanisms that allow bacteria to change their mutation rates. We describe one such mechanism, driven by the action of a mobile element, in the marine bacterium Vibrio splendidus 12B01. We also found similar mobile genetic sequences in the mutS genes of many different bacteria, including clinical and agricultural pathogens. These mobile elements might play an as yet unknown role in the evolution of these important bacteria.
Copyright © 2017 Chu et al.
Figures

Similar articles
-
Characterization of the DNA mismatch repair proteins MutS and MutL in a hypermutator Acinetobacter baumannii.Microb Pathog. 2017 Dec;113:74-84. doi: 10.1016/j.micpath.2017.10.001. Epub 2017 Oct 5. Microb Pathog. 2017. PMID: 28988868
-
A mobile genetic element increases bacterial host fitness by manipulating development.Elife. 2021 Mar 3;10:e65924. doi: 10.7554/eLife.65924. Elife. 2021. PMID: 33655883 Free PMC article.
-
Inactivation of mismatch repair increases the diversity of Vibrio parahaemolyticus.Environ Microbiol. 2009 May;11(5):1254-66. doi: 10.1111/j.1462-2920.2008.01853.x. Epub 2009 Jan 22. Environ Microbiol. 2009. PMID: 19161434
-
Eco-evolutionary Dynamics Linked to Horizontal Gene Transfer in Vibrios.Annu Rev Microbiol. 2018 Sep 8;72:89-110. doi: 10.1146/annurev-micro-090817-062148. Epub 2018 Jun 13. Annu Rev Microbiol. 2018. PMID: 29897833 Review.
-
Small mobile sequences in bacteria display diverse structure/function motifs.Mol Microbiol. 2008 Feb;67(3):475-81. doi: 10.1111/j.1365-2958.2007.06068.x. Epub 2007 Dec 10. Mol Microbiol. 2008. PMID: 18086200 Free PMC article. Review.
Cited by
-
Fine-Scale Structuring of Planktonic Vibrio spp. in the Chinese Marginal Seas.Appl Environ Microbiol. 2022 Dec 13;88(23):e0126222. doi: 10.1128/aem.01262-22. Epub 2022 Nov 8. Appl Environ Microbiol. 2022. PMID: 36346224 Free PMC article.
-
Random sequences rapidly evolve into de novo promoters.Nat Commun. 2018 Apr 18;9(1):1530. doi: 10.1038/s41467-018-04026-w. Nat Commun. 2018. PMID: 29670097 Free PMC article.
-
The chromosomal organization of horizontal gene transfer in bacteria.Nat Commun. 2017 Oct 10;8(1):841. doi: 10.1038/s41467-017-00808-w. Nat Commun. 2017. PMID: 29018197 Free PMC article.
-
Genes and Proteomes Associated With Increased Mutation Frequency and Multidrug Resistance of Naturally Occurring Mismatch Repair-Deficient Salmonella Hypermutators.Front Microbiol. 2020 May 8;11:770. doi: 10.3389/fmicb.2020.00770. eCollection 2020. Front Microbiol. 2020. PMID: 32457709 Free PMC article.
-
Hypermutation-induced in vivo oxidative stress resistance enhances Vibrio cholerae host adaptation.PLoS Pathog. 2018 Oct 30;14(10):e1007413. doi: 10.1371/journal.ppat.1007413. eCollection 2018 Oct. PLoS Pathog. 2018. PMID: 30376582 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
